Review Article | DOI: https://doi.org/10.31579/2835-2882/006
The Role of Host Genetics in the Immune Response to Sars Cov-2 and covid-19 Susceptibility and Severity
1 PhD student (Applied Genetics) at Addis ababa university.
2 Associate professor at Addis ababa university.
*Corresponding Author: Bantayehu Bekele Made, Clinical Psychologist, Shahid Beheshti University of medical sciences, Loghman Hakim hospital, Tehran, Iran.
Citation: Bantayehu Bekele Made, Dereje Beyene (2022). The Role of Host Genetics in the Immune Response to Sars Cov-2 and covid-19 Susceptibility and Severity. Clinical Research and Studies, 1(1) DOI:10.31579/2835-2882/006
Copyright: © 2022 Bantayehu Bekele Made, This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received: 22 September 2022 | Accepted: 07 October 2022 | Published: 13 October 2022
Keywords: sars cov-2; covid-19; host genetics; susceptibility; severity
Abstract
Several seasonal coronaviruses are known to circulate in the human population including alpha coronaviruses HCoV-NL63 and HCoV-229E, and the beta corona viruses HCoV-OC43 and HCoV-HKU1. Moreover, since 2003 three other corona viruses such as SARS-CoV-1: the severe acute respiratory syndrome coronavirus; MERS-CoV: the Middle East respiratory syndrome [MERS] coronavirus; and SARS-CoV-2: the causative agent of COVID-19) have emerged as human pathogens. It become one of the leading pathogens of the latest emerging outbreaks of respiratory disease, representing a serious public health burden worldwide. Studies show that Sars-Cov-2 is more infectious than SARS-CoV this is due to higher binding affinity of RBD to ACE2, less exposed RBD (immune escape), and pre-activation by furin (enhanced viral entry). A wide range of disease severity occurs in the patients’ experiences, from asymptomatic cases, mild infections to serious life threatening conditions requiring admission into the intensive care unit (ICU). Although, it is generally reported that age and co-morbidities contribute significantly to the variations in the clinical outcome of the scourge of COVID-19, a hypothetical question of the possibility of genetic involvement of ACE2, HLA, and TLR genes such as TLR7 and TLR3 in the susceptibility and severity of the disease arose when some unique severe outcomes were seen among young patients with no co-morbidity.
Several seasonal coronaviruses are known to circulate in the human population and generally cause relatively mild respiratory tract infections, including the alpha coronaviruses HCoV-NL63 and HCoV-229E, and the beta corona viruses HCoV-OC43 and HCoV-HKU1, phylogenic evidence show their origin were from bat(NL63 and 229E) or rodent-associated (OC43 and HKU1) coronaviruses (Inna et al., 2020). Furthermore, since 2003 three additional coronaviruses (i.e., SARS-CoV-1: the severe acute respiratory syndrome coronavirus; MERS-CoV: the Middle East respiratory syndrome [MERS] coronavirus; and SARS-CoV-2: the causative agent of COVID-19) have emerged as human pathogens, and each is associated with severe infection) (Inna et al., 2020).
Coronaviruses (CoVs) have become one of the leading pathogens of the latest emerging outbreaks of respiratory disease, representing a serious public health burden worldwide (Inna et al., 2020). A novel coronavirus was identified to play a crucial role in the severe acute respiratory syndrome (SARS) in 2003 (Arnold, 2020). Later, the severe acute respiratory syndrome coronavirus-2 (SARSCoV-2), which produces the disease coronavirus-2019 (COVID-19), has emerged in December 2019. This novel virus appears to be highly contagious and has spread rapidly throughout the world, reaching a pandemic state, with important social and health system costs (Adeboboye et al., 2020). Studies show that SARS-CoV-2 is more infectious than SARS-CoV, which may explain why SARS-CoV-2 has caused a more severe pandemic than SARSCoV. Three mechanisms which have been proposed to potentially play a role in this increased SARS-CoV-2 infectivity are: (1) higher binding affinity of RBD to ACE2, (2) less exposed RBD (immune escape), and (3) pre-activation by furin (enhanced viral entry) (Huang and Wang, 2021). Subsequently, a wide range of clinical outcomes have been observed among individuals diagnosed with COVID-19, ranging from mild respiratory infection to acute respiratory disease and death. Disease severity is disproportionately higher among older adults and individuals with underlying comorbidities, although severe cases of COVID-19 have also been reported among young and healthy individuals (Inna et al., 2020).
Since the start of the current SARS-CoV-2 pandemic, scientists have been puzzling over the factors underlying the inter individual and inter-population differences in disease outcomes (Adeboboye et al., 2020). The resulting clinical manifestation of COVID-19 varied enormously, ranging from mild/asymptomatic illness in 80% of patients to a severe respiratory syndrome in 20%, which further progresses to critical illness requiring ventilation in 5% (Adeboboye et al., 2020). For instance in-vivo studies of the angiotensin-converting enzyme 2 (ACE 2) showed higher expression in the kidneys of male than female patients, which may explain the differences in susceptibility and progression of COVID-19 between male and female patients. Ethnic groups often have higher levels of medical comorbidities and lower socioeconomic status, which may increase their risk of contracting COVID-19 through weak cell-mediated immunity (Koppel J. et al., 2020). A recent study noted that ACE2 expression could vary among Asians (significantly higher) compared to African Americans and Caucasians (Zhao Y. et al., 2020).
Previously, the focuses of most studies were on the clinical characteristics, epidemiology, and genomic characterization of SARS-CoV-2 infection. Only few studies were carried out regarding to the role of host genetics in influencing susceptibility and severity of COVID-19. Individuals in the population harbor single nucleotide polymorphisms (SNPs) across a variety of genes (eg, ACE2, TMPRSS2, HLA, CD147, MIF, IFNG, IL6) that have been implicated in the pathology and immunology of SARS-CoV-2 and other pathogenic coronaviruses (Inna et al., 2020). These and other genetic variants may modulate disease susceptibility, increase or decrease disease severity, alter the variety of symptoms developed, and affect the magnitude and/or quality of the immune responses against SARS-CoV-2 (Inna et al., 2020). The current review aim was to discuss the recent findings regarding the role of host genetics to susceptibility and development of severity to Sars cov-2 infection.
Origin and Biology of Sars Cov-2
Origin of Sars Cov-2
Before first detected in December 2019, COVID 19 was inferred to be present in Hubei province, China, for about a month before (Lytras S. et al., 2021). To understand its origin, it is necessary to go back to 2002. During that period, a novel respiratory coronavirus (SARS-CoV) appeared in Foshan, Guangdong province, China, and spread to 29 countries before public health measures controlled its spread in 2003. The zoonotic origin of SARS-CoV was subsequently linked to live animals available at markets. Further sporadic spill-over events of SARS-CoV from animals took place in Guangzhou, Guangdong, and some researchers working with cultured virus were infected in laboratory accidents (Song et al., 2005), but ultimately SARS-CoV was removed from the human population. Trading of susceptible host animals is an important common theme in the emergence of SARS and COVID-19 (Lytras S. et al., 2021).
After three years of SARS epidemic began, Studies show that horseshoe bats (Rhinolophus) in China were harboring related coronaviruses (Li et al., 2005). SARS-related coronavirus (SARSr-CoV) comprises the Sarbecovirus subgenus of the Betacoronavirus genus. It was inferred that a sarbecovirus circulating in horseshoe bats seeded the progenitor of SARS-CoV in an intermediate animal host, most probably civet cats. Further raccoon dogs and badgers were identified as possible intermediate i.e. to have acted as the conduits of transmission to humans from the horseshoe bat reservoir of SARS-CoV, rather than civet cats being a long-term reservoir host species. On the other way, Wuhan Institute of Virology (WIV) in Hubei was speculated as the source of the pandemic because no SARS-CoV-2 intermediate host has been identified to date and owing to the WIV’s geographic location (Lytras S. et al., 2021).

Wuhan city were the origin of SARS-CoV-2 and located at 1500km away from Yunnan province, from where the closest known naturally occurring sarbecovirus collected from horseshoe bats. How did SARS-CoV-2 arrive in Wuhan? Since its emergence, sampling has revealed that coronaviruses genetically close to SARS-CoV-2 are circulating in horseshoe bats, which are dispersed widely from East to West China, and in Southeast Asia and Japan (Lytras S. et al., 2021). The wide geographic ranges of the potential reservoir hosts—for example, intermediate (R. affinis) or least (R. pusillus) horseshoe bat species, which are known to be infected with sarbecoviruses—indicate that the singular focus on Yunnan is misplaced (Lytras S. et al., 2021). Confirming this assertion, the evolutionarily closest bat sarbecoviruses are estimated to share a common ancestor with SARS-CoV-2 at least 40 years ago (Lytras S. et al., 2021), showing that these Yunnan-collected viruses are highly divergent from the SARS-CoV-2 progenitor. The first of these viruses reported by WIV, RaTG13 (Zhou P. et al., 2020), is certainly too divergent to be the SARS-CoV-2 progenitor, providing key genetic evidence that weakens the “lab-leak” notion. Further, three other sarbecoviruses collected in Yunnan independently of the WIV are now the closest bat coronaviruses to SARS-CoV-2 that have been identified: RmYN02, RpYN06, and PrC31 (Lytras S. et al., 2021).
If different routes of transmission to humans are in place, why is emergence so rare that only two major outbreaks have occurred in the last two decades? Spillover events are not so unusual in locations where more frequent human-animal contacts take place. This is indicated by serology studies showing evidence for SARSr-CoV–specific antibodies in people living in rural locations and even higher rates recorded in people living near bat caves (Lytras S. et al., 2021). Spillover risk will increase with human encroachment into rural areas, resulting from new travel networks around and between urban areas. When a novel virus is then exposed to a densely packed human population, such as in Wuhan city, these spillover events have a much higher chance of resulting in substantial onward spread (Lytras S. et al., 2021).
There are controversial reports that SARS Cov-2 cases in china were traced with imported frozen foods. The emergence of SARS-CoV-2 has properties that are consistent with a natural spillover. Although carriage from a bat cave of a sarbecovirus close enough to SARS-CoV-2 to be the progenitor as a research sample to the WIV is theoretically possible, such a scenario would be extremely unlikely relative to the scale of human-susceptible animal contacts routinely taking place in animal trading. Alternatively, bat guano (feces) is collected for use as fertilizer, again on a much larger scale than irregular research visits to bat caves, consistent with rare but ongoing SARSr-CoV transmissions to humans in rural areas (Lytras S. et al., 2021).
Biology of SARS COV-2
SARS-CoV-2 belongs to the virus order Nidovirales, family Coronaviridae, genus Betacoronavirus (enveloped, positive-sense, singlestranded RNA viruses that are zoonotic) and subgenus Sarbecovirus (WHO, 2020). Transmission in human is primarily through direct contact as well as indirect contact with contaminated objects and respiratory droplet (WHO, 2020). Its incubation period varies from 5 to 6 days and ranges 1-14 days. The disease manifests as a flu-like (respiratory) illness characterized by fever, chills, sore throat, dry cough, expectoration, dyspnoea, fatigue, headache, myalgia or arthralgia, and less commonly hemoptysis, conjunctival congestion and gastrointestinal tract involvement i.e. nausea or vomiting and diarrhoea (WHO, 2020).
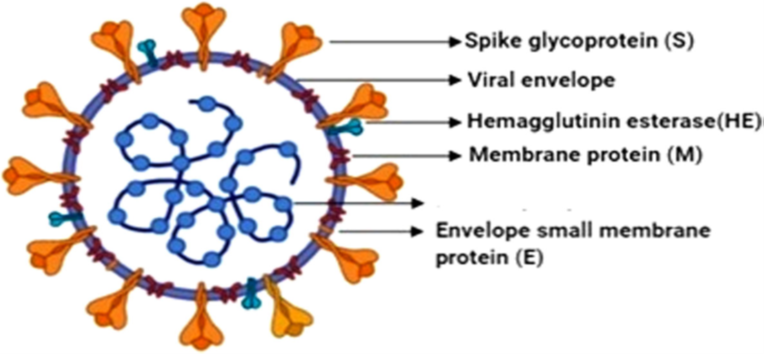
The genome of SARS-CoV-2 is similar to that of other coronaviruses and has four genes that encode the following structural proteins – the S (spike), E (envelope), M (membrane), and N (nucleocapsid) proteins (Fig. 1). The N protein encapsulates the RNA genome (GenBank accession number: MN908947) that measures 29 903 nt in length while the other proteins (the S, E, and M) together comprise the viral envelope (Wu et al., 2021).
Studies show that Cold temperatures favor the survival of cold viruses and reduce immunity in humans and animals by altering the cellular and molecular defenses against pathogens in the upper respiratory tract (Shephard, 1998). On the other hand, reports show, the skewed distribution of COVID-19 mortalities in Italy was not due to the colder weather of the north alone but likely, an interplay of factors including “demographics and health, societal customs and epidemic-specific attitudes, environmental factors and administrative issues” (Goumenou et al., 2020).
Host Genetic Factors in Coronavirus Susceptibility
The spread of the virus has affected all the regions of the world. According to the Worldometery report (September 14, 2021), the global number of cases of COVID-19 has significantly increased, totaling more than 121,716 new cases, while new deaths have increased to over 2,653. This brings the cumulative numbers to over 226,214,149 reported cases and over 4,655,120 deaths globally and in case of Ethiopia 323,715 covid19 cases and 4,967 deaths since the start of the pandemic. The disease has also contributed significantly to the global disease burden, creating a devastating effect on the world economy (Ali et al. 2019).
Fever, fatigue, non-productive cough, decreased leucocyte counts, and radiographic evidence of pneumonia that are very similar to the clinical symptoms of SARS-CoV and MERS-CoV were among the clinical manifestations of Covid-19 (Li et al., 2020). Based on the observations so far, the SARS-CoV-2 causes severe symptoms mainly in aged patients, most especially those with underlying chronic disease conditions. On the other hand, a report from Radboud University Medical Center, Nijmegen, Netherlands notifies two different cases of a pair of previously healthy young brothers with an average age of 26 years from two different families require admission to an intensive care unit (ICU) in rapid succession (Caspar et al., 2020). Further different scenario were reported among health care workers in which they have been severally exposed to the virus remain healthy uninfected while some become infected and die. These unfamiliar cases raise further questions on the consideration of genetic factors and their possible role in compromising the immune system and generally, viral infections have shown certain inter-individual clinical variability. The cause for some people get severe and life-threatening COVID-19, while others are completely asymptomatic or suffer just mild symptoms is worth digging extensively into, most especially with the world searching for answers through research for both therapeutic and prophylactic measures against the virus (Reyfman et al., 2019). The mortality rates were interestingly different between countries. Based on a report of statistica, (2020), the highest mortality rate was seen in European countries such as in the United Kingdom, Italy, and France, reaching 15%. On the contrary, the lowest rate of death from COVID-19 was reported from several Western and South Asian countries such as Singapore (0.06%), Qatar (0.1%), and Bahrain (0.27%) and African countries suchas Rwanda (0.24%) and Uganda (0.42%) (Smatti MK et al., 2021). Studies on host genetic variability towards susceptibility and severity of SARS-Cov-2 so far, focused on ACE-2 and transmembrane protease serine 2 (TMPRSS2). Studies show that TMPRSS2 expression increases ACE2‐mediated invasion of cells by SARS‐CoV‐2, hence the hypothesis that ACE2 and TMPRSS2 variants may modulate viral infectivity in humans, making some individuals more vulnerable than others, seems reasonable. Further, Furin, TMPRSS4, and lyosomal cathepsins shown to be relevant for SARS-CoV-2 entry into host cells (Torre‐Fuentes et al., 2020).
Pathogenesis of SARS-CoV-2 virus
According to Robert (2020), based on the cells that are likely infected, COVID-19 can be classified into three phases that correspond to different clinical stages of the disease. In the initial stage asymptomatic state in 1–2 days of infection the inhaled virus SARS-CoV-2 likely binds to epithelial cells in the nasal cavity and starts replicating. Angiotensin-converting enzyme 2 (ACE2) is the main receptor for both SARS-CoV-1 and 2 (Carter-Timofte et al., 2020). In SARS-CoV-2 infection, cell entry is assisted by the ACE2, which functions together with transmembrane serine protease 2 (TMPRSS2). ACE2 is an important enzyme produced in the renin–angiotensin system (RAS) for a counterbalance action. In the second stage which is next few days the upper airway and conducting airway response and the propagated virus migrates down the respiratory tract along the conducting airways and leads to a more robust innate immune response being triggered possibly through the activation of divers Toll-like receptors (TLRs) (Robert, 2020).
At this time, the disease, COVID-19, becomes clinically manifested. The level of Cys-X-Cys (C-X-C) motif chemokine ligand 10 (CXCL10) and some other innate response cytokine such as interferon (IFN)-alpha, IFN gamma, interleukin (IL)-1β, IL-6, IL-12, and tumor necrosis factor (TNF)-alpha may be predictive of the subsequent clinical course (Reyfman et al. 2019). Epithelial cells that are infected with the virus are a major source of both beta and lambda interferon (Carter et al., 2020). CXCL10 is an interferon responsive gene that has an excellent signal to the alveolar type II cell response to both SARS-CoV and influenza (Hancock et al., 2018, Huang et al., 2020). CXCL10 has also been reported to be useful as a disease marker in SARS (Debnath et al., 2020). In the final stage, hypoxia and progression to acute respiratory distress syndrome (ARDS) are usually seen, characterized by cases of development of pulmonary infiltrates and very severe disease (Robert, 2020).
The major cause of death in most covid-19 patients is acute respiratory distress syndrome (ARDS). Previous survey of 41 SARS-CoV-2- infected patients in admission during the outbreak showed that six of them died from ARDS (Adeboboye et al., 2020). ARDS is majorly experienced as shortness of breath, and it is a common immune pathological event in SARS-CoV and MERS-CoV infections (Mehta et al., 2020). The vital mechanism for the development of ARDS is cytokine storm (Nikolaidis et al., 2017; Schnabel, 2020). Cytokine storm arises from the release of large numbers of pro-inflammatory cytokines (Caspar et al., 2020). This cytokine storm triggers a deleterious attack by the immune system on the body, which in the case of COVID-19 is on the healthy lung tissues of patients. Individuals at older ages are particularly at higher risk because of their diminished immune response and inefficient ability to repair the damaged epithelium. The elderly also have reduced mucociliary clearance, and this may allow the virus to spread to the gas exchange units of the lung more readily (Chen et al., 2020).
Genetic perspective to the determinants of severity of COVID-19
Infections, viral replication, and inflammation is shown to be influenced by host genes and based on their effect on susceptibility and severity viral diseases may classified into categories like virus entry receptors, co-receptors, or receptor-modifying enzymes. Further, Gene polymorphism can influence viral disease severity (Kenney et al., 2017; Carter et al., 2020). It has been observed that genetic mutations or defects in different facets of cellular innate and adaptive immune responses to viral infections associated with enhanced severity of numerous viral infections. So far many of the studies carried out were focus on immunity related genes (Kenney et al., 2017).
A study carried out by Casanova et al. describes the possibility of some inborn errors of immunity which could be either monogenic (single gene) or Mendelian, and that previously healthy, young patients with severe COVID-19 may carry causal genetic variants (Casanova and Abel, 2020). The production of Interferon has major role in controlling mechanism of the immune system in clearing the SARSCov-2. In case of IFN dependent virus control it profoundly impaired during initial infection in patients with early-onset of pneumonia, whereas those whose condition deteriorates later could have milder IFN deficiency or genetically determined excessive inflammation (Casanova and Abel, 2020).
Variation among human leukocyte antigen alleles were also identified as possible genetic determinant of susceptibility and severity. To identify potential susceptibility genes among cohorts afflicted with certain specific clinical manifestations of viral infections GWAS study were used whole exome sequencing (WES) has been adopted to identify candidate gene polymorphisms responsible for specific viral disease phenotype (Kenney et al., 2017; Carter et al., 2020; Casanova and Helen, 2020). A study carried by Smatti MK et al. (2021) based on 74 SNPs, located in 10 genes: ICAM3, IFN-g, CCL2, CCL5, AHSG, MBL, Furin, TMPRSS2, IL4, and CD209 and found Qatari genomes revealed significantly lower AF of risk variants linked to SARS-CoV-2 severity (CCL2, MBL, CCL5, AHSG, and IL4) compared to that of 1000Genome and/or the EAS population (up to 25-fold change). Conversely, SNPs in TMPRSS2, IFN-g, ICAM3, and Furin were more common among Qataris (average 2-fold change). Inter-population analysis showed that the distribution of risk alleles among Europeans differs substantially from Africans and EASs. Remarkably, Africans seem to carry extremely lower frequencies of SARS-CoV-2 susceptibility alleles, reaching to 32-fold decrease compared to other populations (Smatti MK et al., 2021). Another study carried by Li et al. (2021) to reveal genetic determinants of susceptibility to COVID-19 severity in the Chinese population. They found two loci on chromosome 11q23.3 and 11q14.2, were significantly associated with the COVID-19 severity. Understanding such genetic basis of severity to viral infections could project the world further into genetic diagnosis of diseases and infections.
Variations of angiotensin-converting enzyme 2 (ACE2) gene as important gateway for severity of SARS-Cov-2 infection
ACE2 have many physiological roles under its three main function: a negative regulator of the renin-angiotensin system, facilitator of amino acid transport, and the severe acute respiratory syndrome-coronavirus (SARS-CoV) and SARS-CoV-2 receptor. It is mainly expressed in the lungs, cardiovascular system, gut, kidneys, central nervous system, and adipose tissue. Recent studies shown that it serves as SARS-CoV-2 receptor, the infective agent responsible for coronavirus disease 2019, providing a critical link between immunity, inflammation, ACE2, and cardiovascular disease (Mahmoud et al., 2020).
The dynamics of spatial transmission of COVID-19 may be attributed to the variations within ACE2 genes. The X-chromosomes have high number of immune related genes which are responsible for both innate and adaptive immune responses to infection. The ACE2 gene is located on the X- chromosome and is found on the locus Xp22.2. It is 41.04 kb long and contains 18 or 19 exons which exist in two isoforms (Genbank, NT011757) [8]. Over expression of ACE2 might predispose patients to varying degree of severity of this infection as a result of its potential functional variations which have been shown to alter its activities during transcription (Huang et al., 2020).
Cao et al. (2020) identified genetic variants, which may potentially alter ACE2 gene expression. Although it is important to note that while there is a possibility of genetic involvements in the mutual expression levels of ACE and ACE2, scientist have also proposed that the possible cause of these expressions could be based on individual base line health, which further determines which of the two most important angiotensin is expressed in the individual, either angiotensin II or angiotensin 1-7 as seen in the RAS pathway. Young, healthy, and physically fit individual will tend to express more of angiotensin 1-7 which allows for regulation of the pathway and cleavage of ACE2 by enzyme sheddase, leading to less expressions of the ACE2 receptor required for the entry of SAR-CoV-2, unlike in older individuals who have higher risks of developing hypertension, diabetes, heart failure etc., which activates the production of more angiotensin II necessary for higher expression of ACE2. Pathological alterations of the ACE2 pathway seems to cause an increased severity of COVID-19 among patients who suffer from hypertension and diabetes mellitus (DM) as these diseases are both modulated by ACE2 (Debnath et al., 2020)
According to different studies, because of the presence of high ACE2 density in males lung; males are more susceptible to SARS-CoV-2 infection. A study carried out in Asia based on single-cell RNA sequencing (RNA-seq) analysis indicated that Asian males may have a higher expression of tissue ACE2 (Marionneau et al., 2005). This could be due to the presence of ACE2 gene on the X chromosome, with men having only one allele and two in women. Furthermore, estrogen levels have been reported to upregulate ACE2 expression and activity. This creates a double advantage (i.e., two alleles and estrogen upregulation) which may be responsible for the less severity of COVID-19 in females. A report by the Chinese Centre for Disease Control and Prevention (CDCC) showed an overall female mortality of 1.7% of both suspected cases as well as serologically confirmed cases versus 2.8% in males. In serologically confirmed cases, there was a more marked difference of 2.8
References
- Adeboboye C, Oladejo B, Adebolu T (2020) Immunomodulation: a broad perspective for patients’ survival of COVID-19 infection. Eur J Biol Res 10: 217–224
View at Publisher | View at Google Scholar - Albright FS, Orlando P, Pavia AT, Jackson GG, Albright LAC. Evidence for a heritable predisposition to death due to influenza. J Infect Dis. 2008;197:18–24.
View at Publisher | View at Google Scholar - Ali SA, Baloch M, Ahmed N, Ali AA, Iqbal A (2020) The outbreak of coronavirus disease 2019
View at Publisher | View at Google Scholar - Alicia SM (2020) A review of HLA allele and SNP associations with highly prevalent infectious diseases in human populations. Swiss Med Wkly 150: w20214
View at Publisher | View at Google Scholar - Arnold RH (2020) COVID-19 – does this disease kill due to imbalance of the renin angiotensin system (RAS) caused by genetic and gender differences in the response to viral ACE 2 attack? Heart Lung Circ 29(7):964–972
View at Publisher | View at Google Scholar - Britanova OV, SHugay M, Merzlyak EM, Staroverov DB, Putintseva EV, Turchabinova MA et al
View at Publisher | View at Google Scholar - Cao Y, Li L, Feng Z, Wan S, Huang P, Sun X et al (2020) Comparative genetic analysis of the novel corona-virus (2019-nCoV/SARS-CoV-2) receptor ACE2 in different populations. Cell Discov 6:11
View at Publisher | View at Google Scholar - Carter-Timofte ME, Jørgensen SE, Freytag ME, Thomsen MM, Andersen NB, Al-Mousawi A et al (2020) Deciphering the role of host genetics in susceptibility to severe. COVID 19 1606(11):1–14
View at Publisher | View at Google Scholar - Casanova JL, Abel L (2020) Lethal infectious diseases as inborn errors of immunity: toward a synthesis of the germ and genetic theories. Annu Rev Pathol 1:l031920–l101429
View at Publisher | View at Google Scholar - Casanova JL, Helen CS, the COVID Human Genetic Effort (2020) A global effort to define the human genetics of protective immunity to SARS-CoV-2 infection. Cell 181:1194–1199.
View at Publisher | View at Google Scholar - Caspar I, Van der Made MD, Janneke SH et al (2020) Presence of genetic variants among young men with severe COVID-19. JAMA. 324(7):1. https:// doi.org/10.1001/jama.2020.13719.
View at Publisher | View at Google Scholar - Chen N, Zhou M, Dong X et al (2020) Epidemiological and clinical characteristics of 99 cases of
View at Publisher | View at Google Scholar - Ciaglia E, Vecchione C, Puca AA (2020) COVID-19 infection and circulating ACE2 levels: protective role in women and children. Front Pediatr 8:206.
View at Publisher | View at Google Scholar - Debnath M, Banerjee M, Berk M (2020) Genetic gateways to COVID-19 infection: implications for risk, severity, and outcomes. The FASEB J 00:1–9
View at Publisher | View at Google Scholar - Fauci AS, Morens DM. The perpetual challenge of infectious diseases. N Engl J Med. 2012;366:454–461.
View at Publisher | View at Google Scholar - Gemmati D, Bramanti B, Serino ML, Secchiero P, Zauli G, Tisato V (2020) COVID-19 and individual genetic susceptibility/receptivity: role of ACE1/ ACE2 genes, immunity, inflammation and coagulation. Might the double Xchromosome in females be protective
View at Publisher | View at Google Scholar - Goumenou M, Sarigiannis D, Tsatsakis A, Anesti O, Docea AO, Petrakis D, et al. COVID-19 in
View at Publisher | View at Google Scholar - Guillot L, Le Goffic R, Bloch S, Escriou N, Akira S, Chignard M et al (2005) Involvement of toll-
View at Publisher | View at Google Scholar - Hancock AS, Stairiker CJ, Boesteanu AC et al (2018) Transcriptome analysis of infected and
View at Publisher | View at Google Scholar - Huang C, Wang Y, Li X et al (2020) Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395(10223):497–506
View at Publisher | View at Google Scholar - Huang, S.-W.;Wang, S.-F (2021). SARS-CoV-2 Entry Related Viral and Host Genetic Variations: Implications on COVID-19 Severity, Immune Escape, and Infectivity. Int. J. Mol. Sci. 2021, 22, 3060. https://doi.org/10.3390/ijms22063060
View at Publisher | View at Google Scholar - Inna G. Ovsyannikova, Iana H. Haralambieva, Stephen N. Crooke, Gregory A. Poland Richard B. Kennedy (2020). The role of host genetics in the immune response to SARSCoV- 2 and COVID-19 susceptibility and severity. Mayo Clinic Vaccine Research Group, Mayo
View at Publisher | View at Google Scholar - Ioannidis I, Ye F, McNally B, Willette M, Flano E (2013) Toll-like receptor expression and induction of type I and type III interferons in primary airway epithelial cells. J Virol
View at Publisher | View at Google Scholar - Iqbal MS, Sardar N, Akmal W, Sultan R, Abdullah H, Qindeel M, Dhama K, Bila M (2020) Role
View at Publisher | View at Google Scholar - Jenifer MB, Sarra EJ, David B (2009) HLA and infectious diseases. Clin Microbiol Rev
View at Publisher | View at Google Scholar - Jonathan Kopel1, Abhilash Perisetti, Ali Roghani, Muhammad Aziz, Mahesh Gajendran and
View at Publisher | View at Google Scholar - Jose JR, Manuel A (2020) COVID-19 cytokine storm: the interplay between inflammation and coagulation. Lancet Respir. Med S2213-2600(20):30216–30212
View at Publisher | View at Google Scholar - Kenney AD, Dowdle JA, Bozzacco L, McMichael TM, Gelais C, Panfil AR et al (2017) Human genetic determinants of viral diseases. Annu Rev Genet 51: 241–263.
View at Publisher | View at Google Scholar - Li Nie, Shi-Yu Cai, Jian-Zhong Shao and Jiong Chen (2018). Toll-Like Receptors, Associated Biological Roles, and Signaling Networks in Non-Mammals.doi.org/10.3389/fimmu.2018.01523
View at Publisher | View at Google Scholar - Li W, Shi Z, Yu M et al., (2005). Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005 Oct 28;310(5748):676-9. doi: 10.1126/science.1118391.
View at Publisher | View at Google Scholar - Li X, Geng M, Peng Y, Meng L, Lu S (2020) Molecular immune pathogenesis and diagnosis of COVID-19. J Pharm Anal 10(2):102. https://doi.org/10.1016/j. jpha.2020.03.001.
View at Publisher | View at Google Scholar - Li, Y., Ke, Y., Xia, X., Wang, Y., Cheng, F., & Liu, X. et al. (2021). Genome-wide association study of COVID-19 severity among the Chinese population. Retrieved 20 September 2021.
View at Publisher | View at Google Scholar - Lin M, Tseng HK, Trejaut JA, Lin H, Loo J, Chu C et al (2003) Association of HLA class I with
View at Publisher | View at Google Scholar - Lucas G, John B, Houda A (2020) Deciphering the TCR repertoire to solve the COVID-19 mystery. Trends Pharmacol Sci 41(8):518–530
View at Publisher | View at Google Scholar - Mahmoud Gheblawi, Kaiming Wang, Anissa Viveiros, Quynh Nguyen, Jiu-Chang Zhong, Anthony J. Turner, Mohan K. Raizada, Maria B. Grant, and Gavin Y. Oudit (2020). Angiotensin-Converting Enzyme 2: SARS-CoV-2 Receptor and Regulator of the Renin-
View at Publisher | View at Google Scholar - Marionneau S, Airaud F, Bovin NV, Le Pendu J, Ruvoën-Clouet N (2005) Influence of the combined ABO, FUT2 and FUT3 polymorphism on susceptibility to Norwalk virus attachment J. Infect Dis Ther 192:1071–1077.
View at Publisher | View at Google Scholar - Mehta P, McAuley DF, Brown M, Sanchez E, Tattersall RS, Mansion JJ (2020) COVID-19: consider cytokine storm syndrome and immunosuppression. Lancet 395(10229):1033–2034
View at Publisher | View at Google Scholar - Netea MG, Wijmenga C, O'Neill LA (2012) Genetic variation in toll-like receptors and disease susceptibility. Nat Immunol 13(6):535–542
View at Publisher | View at Google Scholar - Nguyen A, David JK, Maden SK et al (2020) Human leukocyte antigen susceptibility map for SARS-CoV-2. Am Soc Microbiol 1128:00510
View at Publisher | View at Google Scholar - Nikolaidis NM, Noel JG, Pitstick LB et al (2017) Mitogenic stimulation accelerates influenza-induced mortality by increasing susceptibility of alveolar type II cells to infection. Proc Natl Acad Sci U S A 114:E6613–E6622
View at Publisher | View at Google Scholar - Reyfman PA, Walter JM, Joshi N et al (2019) Single-cell transcriptomic analysis of human lung
View at Publisher | View at Google Scholar - Robert J. Mason (2020). Pathogenesis of COVID-19 from a cell biology perspective. National Jewish Health, Dept of Medicine, Denver, CO, USA. 7 doi.org/10.1183/13993003.00607-2020.
View at Publisher | View at Google Scholar - Schnabel J (2020) Why is COVID-19 mild for some, deadly for others? Cornell chronicles. Toggle
View at Publisher | View at Google Scholar - Shephard RJ. Immune changes induced by exercise in an adverse environment. Can J Physiol Pharmacol. 1998;76:539–546.
View at Publisher | View at Google Scholar - Smatti MK, Al-Sarraj YA, Albagha O and Yassine HM (2020). Host Genetic Variants Potentially Associated With SARS-CoV-2: A Multi-Population Analysis. Front. Genet. 11:578523. doi: 10.3389/fgene.2020.578523
View at Publisher | View at Google Scholar - Song HD, Tu CC, Zhang GW, Wang SY et al., (2005). Cross-host evolution of severe acute respiratory syndrome coronavirus in palm civet and human. Proc Natl Acad Sci U S A.
View at Publisher | View at Google Scholar - Sørensen TIA, Nielsen GG, Andersen PK, Teasdale TW. Genetic and environmental influences on premature death in adult adoptees. N Engl J Med. 1988;318:727–32.
View at Publisher | View at Google Scholar - Spyros Lytras, wei Xia, joseph Hughes, xiaowei Jiang, and David L. Robertson (2021) The animal
View at Publisher | View at Google Scholar - Torre‐Fuentes, L., Matías‐Guiu, J., Hernández‐Lorenzo, L., Montero‐Escribano, P., Pytel, V., Porta‐Etessam, J., Gómez‐Pinedo, U., & Matías‐Guiu, J. A. (2020). ACE2, TMPRSS2 , and Furin variants and SARS‐CoV‐2 infection in Madrid, Spain. Journal of Medical Virology, 93(2), 863–869. https://doi.org/10.1002/jmv.26319
View at Publisher | View at Google Scholar - World Health Organization (WHO). Report of the WHO-China Joint Mission on Coronavirus Disease 2019 (COVID-19), 2020. https://www.who.int/docs/default-source/coronaviruse/who-china-joint-mission-on-covid-19-finalreport.pdf. Accessed 12
View at Publisher | View at Google Scholar - Wu C, Liu Y, Yang Y, Zhang P, Zhong W, Wang Y, et al. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm Sin B.2020. https://doi.org/10.1016/j.apsb.2020.02.008 In Press. Accessed 12 July 2021.
View at Publisher | View at Google Scholar - Xu ZL, Shi L, Wang Y et al (2020) Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med 8(4):420–422
View at Publisher | View at Google Scholar - Zhao Y, Zhao Z, Wang Y, Zhou Y, Ma Y, Zuo W. Single-cell RNA expression profiling of ACE2,
View at Publisher | View at Google Scholar - Zhou, P., Yang, XL., Wang, XG. et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273 (2020). https://doi.org/10.1038/s41586-020-2012-7.
View at Publisher | View at Google Scholar

 Clinic
Clinic
