Research Article | DOI: https://doi.org/10.31579/2834-8486/029
Structural Peptidomics of Diabetic Saliva Reveals Mucosal Barrier Remodeling and Design Cues for Biofabrication
- Rui Miguel Pinheiro Vitorino *
Cardiovascular R&D Centre – UnIC@RISE, Department of Surgery and Physiology, Faculty of Medicine of the University of Porto, Porto, Portugal.
*Corresponding Author: Rui Miguel Pinheiro Vitorino, Cardiovascular R&D Centre – UnIC@RISE, Department of Surgery and Physiology, Faculty of Medicine of the University of Porto, Porto, Portugal.
Citation: Rui Miguel Pinheiro Vitorino, (2024), Structural Peptidomics of Diabetic Saliva Reveals Mucosal Barrier Remodeling and Design Cues for Biofabrication, Biomedical and Clinical Research. 4(2); DOI: 10.31579/2834-8486/029
Copyright: © 2024, Rui Miguel Pinheiro Vitorino. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Received: 21 February 2025 | Accepted: 07 March 2025 | Published: 27 March 2025
Keywords: saliva; peptidomics; type 2 diabetes; AlphaFold; bioprinting; mucosal barrier
Abstract
Saliva provides a non-invasive window into systemic health; however, most diabetes-related omics research focuses on whole proteins rather than the structural information contained in endogenous peptides. The human salivary peptidome was characterised in individuals with type 2 diabetes (T2D) compared to normoglycaemic controls using a peptide-centred, library-free DIA-NN approach. A total of 199 significantly modulated peptides (|FC| ≥ 1.5; p < 0.05) were identified, showing a marked asymmetry pattern with increased levels of apolipoprotein, complement, mucin, and haemoglobin fragments in T2D. These changes reflect coordinated adjustments in mucosal barrier regeneration, lipid metabolism, and microvascular permeability. AlphaFold modelling of representative peptides revealed distinct structural motifs – flexible mucin loops, amphipathic helices, and redox-active globin segments – suggesting roles in hydration, immune regulation, and hydrogel behaviour. These results show that salivary peptides are structurally and functionally relevant indicators that surpass conventional protein-level analysis. This study combines deep peptidomics with structure prediction to clarify molecular changes in the diabetic salivary environment and establishes a framework for using endogenous peptides as both diagnostics and functional components for mucosa-inspired bioinks in tissue engineering
Introduction
Type 2 diabetes (T2D) is a systemic metabolic condition characterised by persistent hyperglycaemia, mild inflammation, and progressive vascular and epithelial impairment. While blood-based biomarkers remain the clinical standard for assessing metabolic health, there is growing interest in saliva as a non-invasive diagnostic medium that can reflect both local and systemic biochemical changes [1]. Saliva captures the contributions of salivary glands, gingival crevicular fluid, mucosal surfaces, and microvasculature, serving as a sensitive indicator of inflammatory status, epithelial integrity, and metabolic stress. Consequently, salivary proteomics has emerged as a valuable adjunct for monitoring metabolic diseases, particularly in contexts requiring frequent sampling or patient-friendly collection methods [2,3].
Recent metaproteomic studies have shown that the salivary proteome of individuals with Type 2 diabetes is altered at multiple levels, including changes in immune-related proteins, mucins, and microbial components. Protein-level analyses provide only a general overview of these changes and often obscure more subtle molecular variations occurring at the peptide level [3,4]. Peptide-centric approaches, especially when combined with modern data-independent acquisition (DIA) and neural network-assisted processing, enable more accurate quantification of small fragments that directly indicate proteolytic activity, barrier integrity, redox balance, and tissue leakage. These peptides often serve as the earliest molecular indicators of diseased tissue remodelling, yet they remain largely unexplored in the context of T2D saliva [5,6].
Moreover, understanding not only which peptides are altered but also how their structural features may influence biological behaviour is increasingly relevant for fields such as biofabrication and mucosa-mimetic bioprinting [7]. The mechanical and biochemical properties of bioinks, such as hydration, lubrication, diffusivity, immunological compatibility, and redox stability, are significantly affected by the peptides and proteins present in the oral environment. Structural modelling techniques, such as AlphaFold, now allow the correlation of peptide abundance changes with predicted physicochemical patterns, providing greater insight into how disease-related secretions may interact with engineered tissues [8–10].
This study uses a high-resolution, peptide-focused DIA-NN approach to analyse the salivary peptidome in individuals with Type 2 diabetes and to identify the most distinctive human peptide fragments. By integrating quantitative peptidomics with AlphaFold structural predictions, we provide both biochemical and structural perspectives on saliva alterations associated with T2D. The findings advance the current understanding of the diabetic salivary phenotype and highlight peptide features that may be important for mucosal biology and oral-tissue bioprinting, thereby creating opportunities for diagnostic and biomaterials-related applications.
2.Materials and Methods
2.1. Saliva Collection and Sample Preparation
Saliva samples were processed following the methodology described by Samodova et al. (2025) [11], with minor modifications for peptide-focused analysis. In brief, 20 µL of saliva was mixed with 10 µL of lysis buffer containing 1% sodium deoxycholate (SDC), 100 mM Tris-HCl (pH 8.5), 10 mM TCEP, and 40 mM chloroacetamide. The mixture was heated at 99 °C for 10 minutes to denature proteins and disrupt secondary structures, then subjected to 4 minutes of probe sonication to ensure complete homogenisation. Proteins underwent a standard two-step digestion protocol. Samples were first incubated with Lys-C (1:250, 1 hour, 37 °C), followed by overnight digestion with trypsin (1:100, 37 °C), as per the referenced protocol. Digestion was stopped by adding 10% TFA (1:10, v/v), and peptides were clarified by centrifugation (17,000 × g, 5 minutes). Supernatants were quantified by spectrophotometry and desalted using C18 StageTips. Approximately 200 ng of purified peptides per sample were loaded onto Evotips for LC–MS/MS analysis, following the preparation protocol described in the parent document.
2.2. Liquid Chromatography–Mass Spectrometry (LC–MS/MS)
Peptide separation and analysis were performed using an Evosep One liquid chromatography system coupled with a Bruker timsTOF instrument. Samples were analysed using the 21-minute “60 samples per day” protocol developed by Samodova et al., enabling a high-throughput and comprehensive proteomic workflow. Chromatographic separation was achieved with an 8-cm PepSep C18 column (150 µm inner diameter; 1.5 µm particles), and ionisation was facilitated by a CaptiveSpray source.
Data were acquired using DIA-PASEF with the following parameters:
• m/z acquisition range: 400–1200
• TIMS ion mobility range: 0.6–1.43 Vs/cm²
• Ramp/accumulation duration: 100 ms
DIA scheme: 32 × 25 Da windows, each acquired through 2 × 16-scan repetitions.
• Total cycle duration: approximately 1.8 seconds
This acquisition strategy combines ion mobility separation with wide-window DIA to achieve comprehensive peptide coverage within a short gradient.
2.3. Proteomic Data Processing (DIA-NN Re-Analysis)
Raw DIA-PASEF files were reanalysed with DIA-NN (v.1.8+) in library-free mode to improve peptide-level resolution. Database searches were performed using a hybrid FASTA that includes the reviewed (Swiss-Prot) proteome of Homo sapiens.
Peptide and Precursor Parameters: minimum peptide length, 5 amino acids; maximum peptide length, 40 amino acids; precursor mass range, 500–1800 Daltons; maximum allowable missed cleavages, 2; enzymatic specificity, fully tryptic; fixed modification, carbamidomethylation of cysteine; variable modifications, oxidation of methionine and acetylation at the N-terminus.
FDR and Normalisation: FDR threshold, 1% at both precursor and protein levels; interference correction, enabled; RT-dependent normalisation, enabled.
Quantification was performed at the precursor level using the neural network scoring model of DIA-NN.
2.4. Peptide-Centric Statistical Analysis (MetaboAnalyst 6.0)
The precursor intensities from DIA-NN were imported into MetaboAnalyst 6.0 for statistical analysis. Data were analysed using log₂ transformation, filtering to retain features present in more than 70% of samples within at least one group, and imputation of missing values with the K-nearest neighbours (KNN) method. Analyses included Principal Component Analysis (PCA) and volcano plot analysis integrating fold-change (|FC| ≥ 1.5) and t-tests, with a significance threshold of p < 0>
2.5. AlphaFold Structural Modelling of Differentially Abundant Peptides
To determine whether peptides with differential abundance in T2D saliva displayed structural features relevant to bioink behaviour, such as amphipathic helices, flexible regions, or accessible hydrophobic surfaces, a representative set of peptides was modelled using AlphaFold/ColabFold. Peptides were selected based on biological significance, effect size, and potential influence on lubrication, matrix interaction, redox balance, or immunomodulation in bioprinted constructs. Structural predictions were generated using standard AlphaFold/ColabFold settings, yielding three- dimensional conformations with per-residue pLDDT confidence scores. Models were evaluated for secondary structure elements, flexibility, charge distribution, and solvent-exposed motifs relevant to bioprinting applications. The final models were presented in both ribbon and surface representations for comparative analysis.
3. Results
3.1. Differential Salivary Peptidome in T2D vs. Controls
A peptide-focused DIA-NN study revealed distinct alterations in the salivary peptidome of patients with type 2 diabetes (T2D) compared to normoglycaemic controls. A total of 199 peptides met the significance criteria (|FC| ≥ 1.5; p < 0 xss=removed>
The most significantly altered features included haemoglobin-derived peptides (P69905, P68871, P02042), with fold-changes between 4 and nearly 8. These elevations likely reflect microvascular fragility or subclinical gingival bleeding, common complications in diabetes, providing biologically consistent justification for their marked increase in saliva. Multivariate analysis supported this global restructuring: principal component analysis showed clear separation between T2D and control participants, indicating that the combined peptide signature defines a coherent and disease-specific salivary phenotype.
Collectively, these findings indicate a dual pattern in diabetic saliva: a specific decrease in structural and regulatory peptides, alongside a widespread increase in complement-related, acute- phase, lipoprotein-derived, and haemoglobin fragments. This composite signature illustrates the interconnected effects of low-grade inflammation, epithelial barrier remodelling, and increased vascular permeability typical of metabolic dysregulation. The extent and specificity of these peptide changes highlight their potential as non-invasive biomarkers for identifying or monitoring physiological changes associated with T2D.
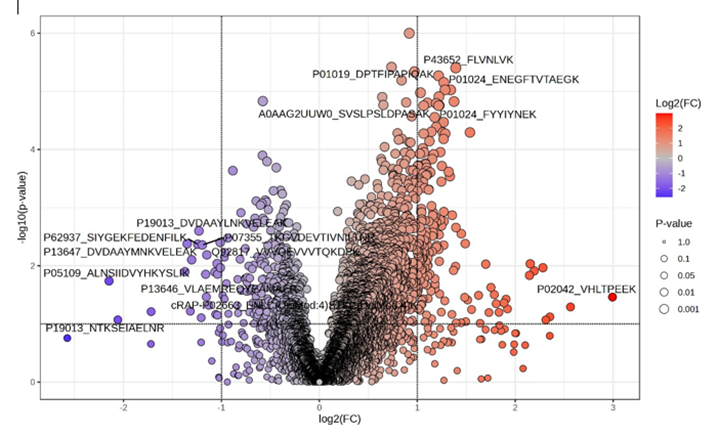
Figure 1. Volcano plot showing differentially abundant salivary peptides in T2D versus normoglycemic controls. Each point represents a quantified peptide, plotted according to its log₂ fold change (x-axis) and – log₁₀(p-value) (y-axis). Vertical dashed lines indicate the fold-change threshold (|FC| > 1.5), while the horizontal dashed line marks the significance cutoff (p < 0>
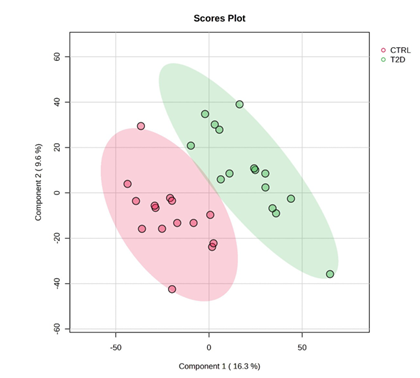
Figure 2. Principal Component Analysis (PCA) of the salivary peptidome distinguishing T2D and control groups. Scores plot of the first two principal components (PC1: 16.3% variance; PC2: 9.6% variance) derived from peptide-level DIA-NN intensities. Each point represents a saliva sample, categorised by colour (red: controls; green: T2D). Ellipses indicate the 95% confidence intervals for each cohort. The substantial separation between groups across both components indicates consistent and disease-specific differences in the salivary peptide profile associated with type 2 diabetes.
Scheme S1. Enhanced annotation of differentially abundant salivary peptides in T2D versus controls.
This table lists all peptides identified as significantly modulated between individuals with type 2 diabetes (T2D) and normoglycemic controls based on DIA-NN precursor quantification (|FC| ≥ 1.5; p < 0>
3.2. AlphaFold Structural Modelling
AlphaFold modelling of differentially abundant peptides provided a structural perspective that enhances the quantitative analysis of salivary peptidomics. Although brief, the selected peptides displayed unique physicochemical characteristics that clarify how changes in peptide abundance associated with T2D may influence lubrication, immunological signalling, and matrix dynamics in a bioprinting context. The two peptides derived from MUC5B, IVTENIPCGTTGTTCSK and TGLLVEQSGDYIK, showed the greatest flexibility and extension in their conformations. The extended region, containing two cysteine residues and multiple glycine/threonine repeats, was expected to adopt an open, highly dynamic conformation, consistent with the disordered segments that confer elasticity and hydration to mucin polymers. The second MUC5B peptide displayed a short turn-like structure with exposed polar groups capable of interacting with both aqueous and hydrogel environments. Together, these features highlight that the increased presence of MUC5B peptides in T2D saliva represents not only a compositional change but also a shift in structural motifs essential for mucus viscosity and epithelial barrier function.
In contrast, peptides from ApoA1 and ApoA2 demonstrated distinct amphipathic properties. AlphaFold predicted a partially helical conformation for the ApoA1 fragment DSGRDYVSQFEGSALGK, with hydrophobic residues aligned along one face of the helix and acidic or polar residues grouped on the opposite side. The shorter ApoA2 peptide SPELQAEAK exhibited a compact helix-turn motif with a similar arrangement of charged and nonpolar residues. The amphipathic structures typical of lipid-binding proteins suggest that the elevated levels of apolipoprotein peptides in T2D saliva may influence interfacial tension, micelle-like behaviour, and polymer–protein interactions in mucosa-mimetic bioinks. Peptides associated with complement and protease-inhibitor pathways, A2M (TEHPFTVEEFVLPK), C3 (TIYTPGSTVLYR), and C4BP (EDVYVVGTVLR)—displayed greater structural compactness and stability. AlphaFold predicted short, structured motifs with defined regions of hydrophobic and charged residues, resulting in compact interaction surfaces rather than extended flexible regions. These features support transient binding events and may regulate protease inhibition, complement signalling, or cell–matrix communication when incorporated into printed tissue. Their enrichment in T2D saliva thus introduces an immunological dimension to the biochemical environment that bioinks would encounter in vivo. Peptides derived from haemoglobin, particularly the significantly upregulated sequences VLGAFSDGLAHLDNLK and VHLTPEEK, exhibited the most globular and helix-rich conformations. These structures resemble segments of the globin fold and present surface-exposed hydrophobic and polar groups that can participate in redox processes or interact with oxygen and reactive molecules. Their marked increase in T2D saliva, along with their consistent secondary-structure patterns, suggests they may locally affect redox balance or oxygen diffusion, factors known to influence hydrogel stability, cell viability, and tissue remodelling after bioprinting.
The AlphaFold predictions collectively show that the peptides significantly altered in T2D saliva are not random fragments but belong to specific structural categories, flexible mucin loops, amphipathic apolipoprotein helices, compact immune-regulatory motifs, and helix-rich haemoglobin segments, each with functional implications for lubrication, immune signalling, matrix interaction, and redox behaviour. Integrating these structural features with the observed fold-change patterns establishes a mechanistic link between metabolic dysregulation and the physicochemical environment relevant for developing more physiologically representative oral-tissue bioinks.
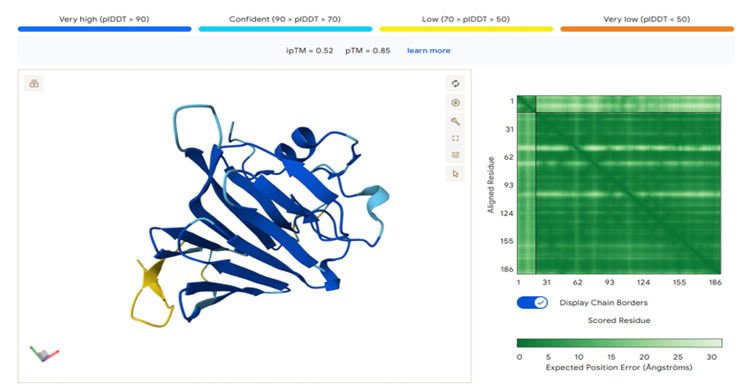
Figure 3. Structures of upregulated MUC5B peptides in T2D saliva predicted by AlphaFold. Ribbon and surface representations of (A) IVTENIPCGTTGTTCSK and (B) TGLLVEQSGDYIK, emphasizing anticipated secondary- structure components and per-residue confidence (pLDDT). The peptide IVTENIPCGTTGTTCSK exhibits an elongated, pliable conformation, with two cysteine residues arranged to facilitate possible intrachain or interchain interactions, aligning with mucin-like polymer characteristics. TGLLVEQSGDYIK exhibits a compact turn motif characterized by flexible termini and a mixed hydrophobic/polar distribution, suggesting potential functions in hydration, lubrication, or polymer interfacing within bioink environments.
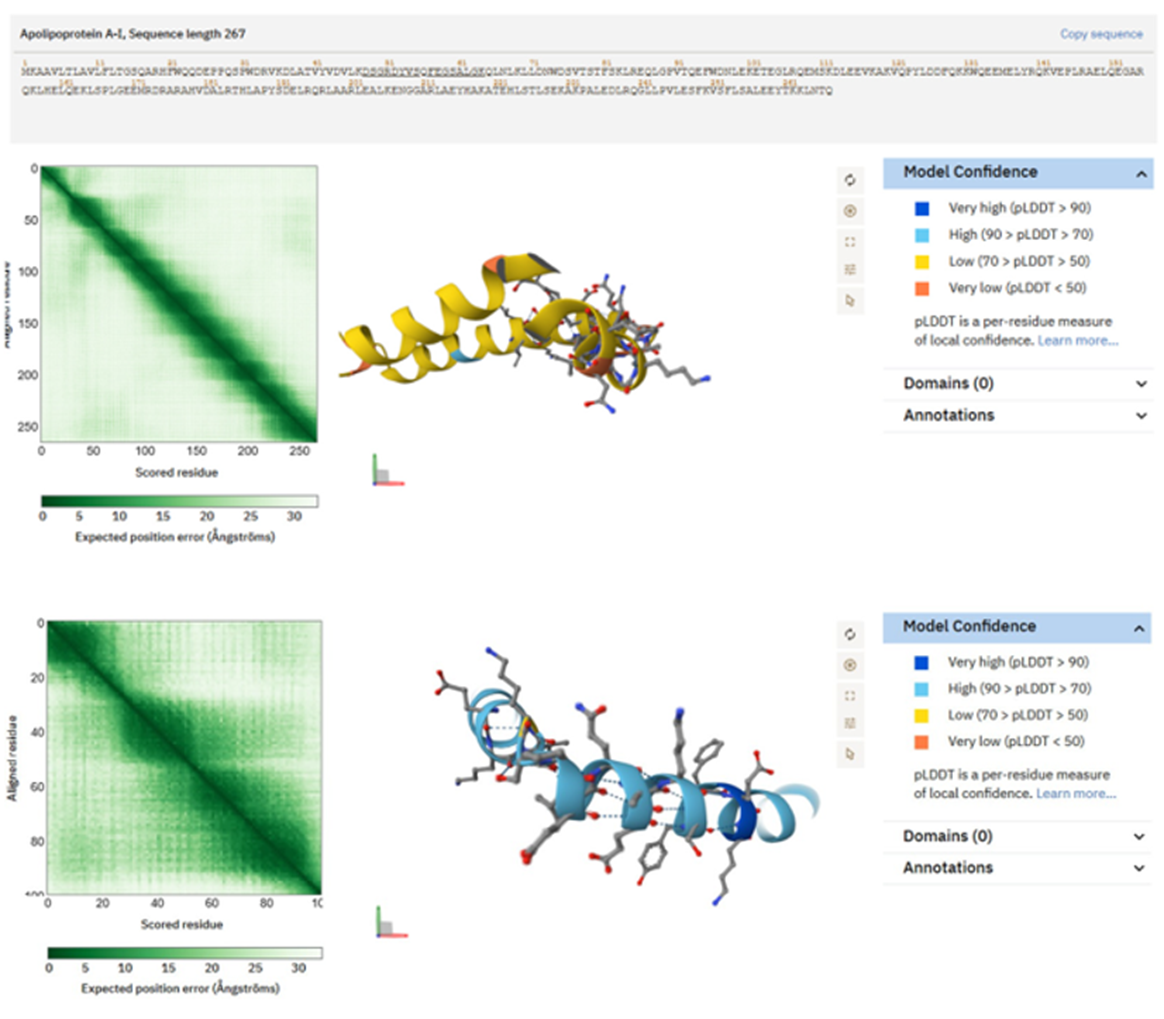
Figure 4. Structures predicted by AlphaFold of peptides produced from ApoA1 and ApoA2, abundant in saliva of individuals with Type 2 Diabetes. Ribbon and surface representations of the ApoA1 peptide DSGRDYVSQFEGSALGK, illustrating a partly helical amphipathic structure with segregation of hydrophobic and polar residues, typical of lipid-associated domains. Structural prediction of the ApoA2 peptide SPELQAEAK, which displays a brief helical/turn motif characterized by an equitable distribution of charged and hydrophobic residues. These characteristics facilitate possible interactions with polymeric matrices, micelle- like environments, or hydrogel constituents pertinent to bioink efficacy.
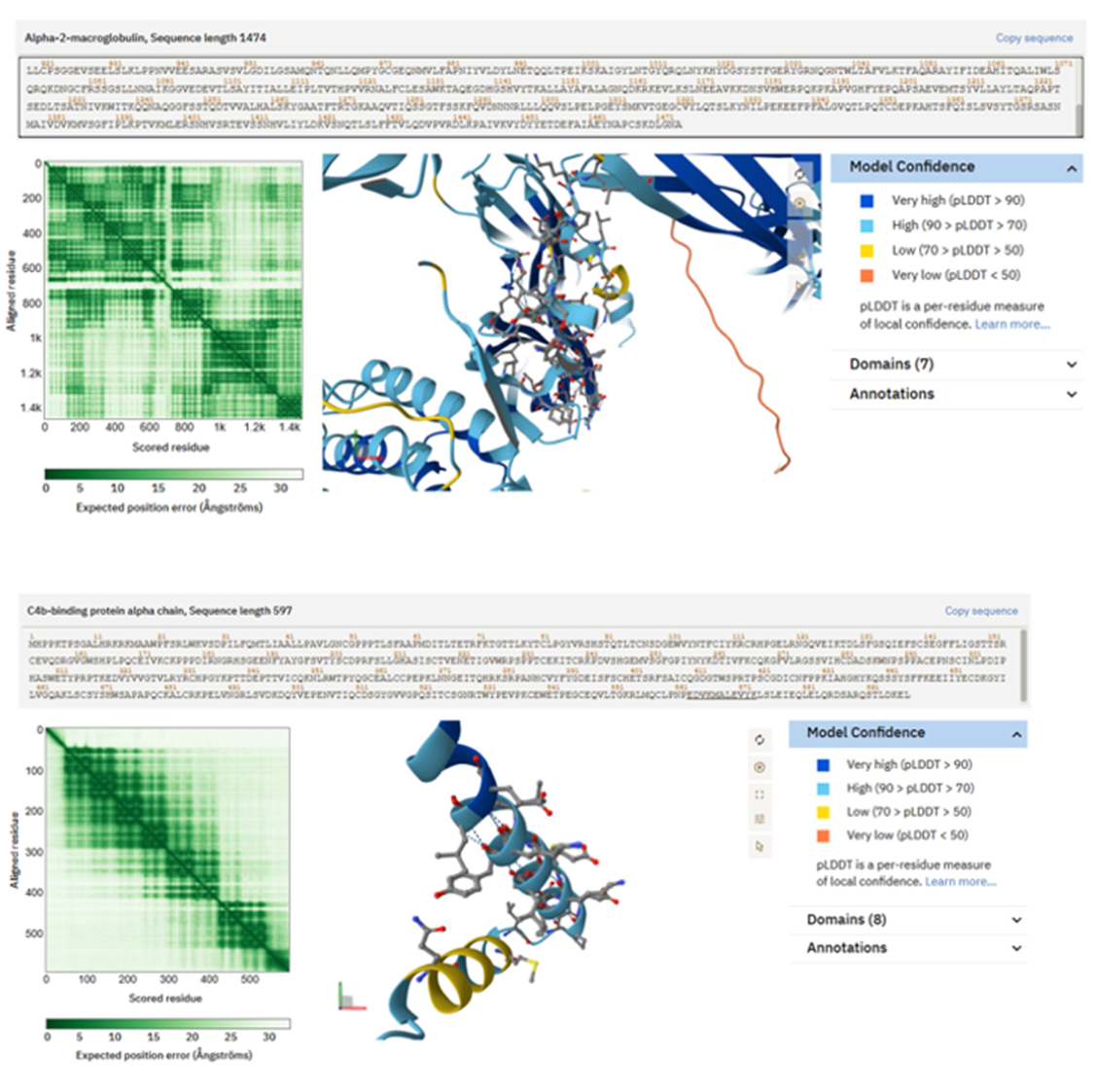
Figure 5. AlphaFold structural predictions for peptides produced from complement and protease inhibitors, abundant in saliva from individuals with Type 2 Diabetes.
Peptide TEHPFTVEEFVLPK from α2-Macroglobulin exhibits an acidic, partly folded motif suitable for interactions with proteases and cytokines. Complement C3 peptide TIYTPGSTVLYR assumes a compact shape, with a combination of aromatic and polar residues creating a possible regulatory interface. The peptide EDVYVVGTVLR from the C4b-binding protein α-chain exhibits hydrophobic clustering encircled by charged residues, indicating potential for scaffold anchoring or surface binding. These structural features elucidate how complement fragments may influence immunological or matrix interactions inside bioprinted tissues.
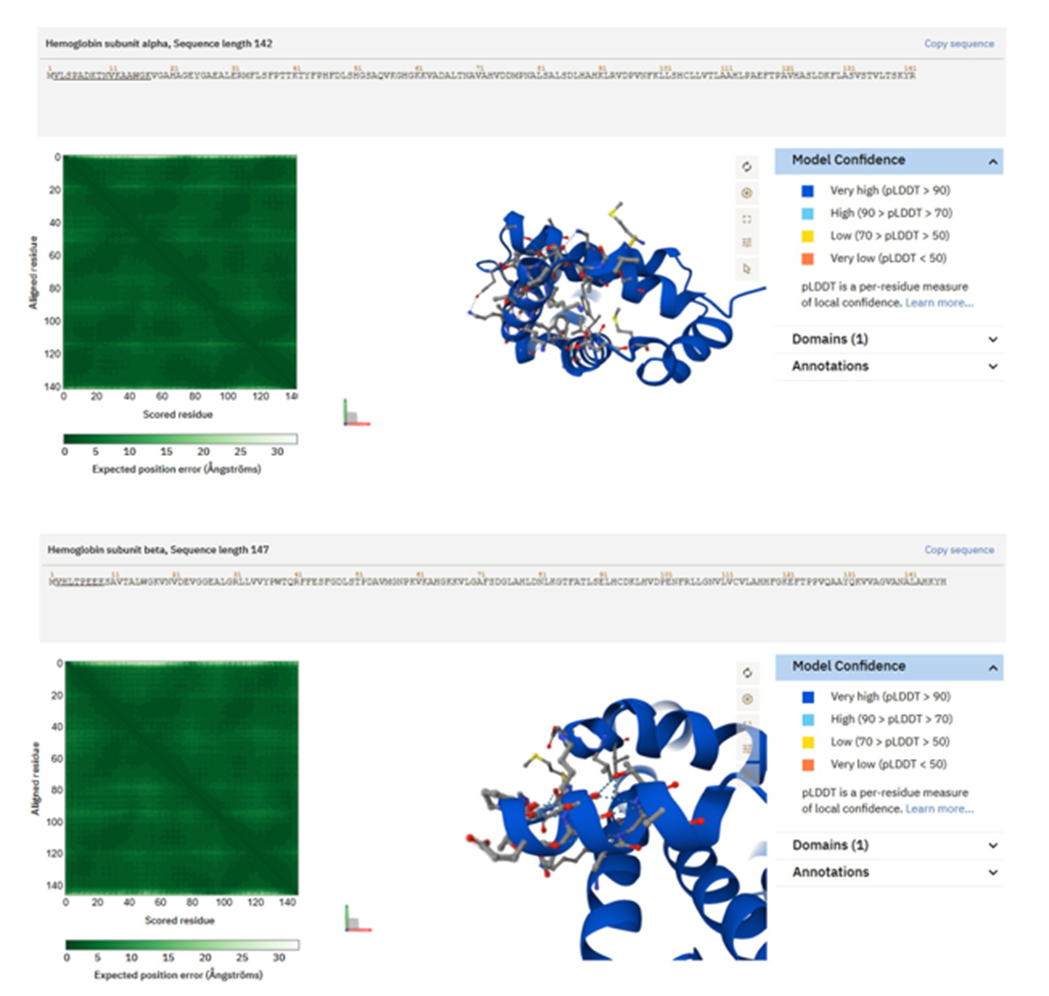
</>< style="text-align:justify;">
Figure 6. Structures of hemoglobin-derived peptides predicted by AlphaFold, abundant in saliva from individuals with Type 2 Diabetes. Hb δ/β-like peptide VLGAFSDGLAHLDNLK exhibits a partly helical conformation typical of globin substructures, characterized by alternating hydrophobic and polar residues that create a structured contact surface. (B) The Hb β-chain peptide VHLTPEEK demonstrates a compact shape characterized by a short protruding helix and a heterogeneous distribution of hydrophobic and charged residues. These structural characteristics may facilitate redox regulation, oxygen management, or matrix interactions within bioprinting contexts.
4.Discussion
This study provides an improved perspective on how type 2 diabetes (T2D) alters the oral environment by focusing on endogenous peptide fragments rather than intact proteins, which more accurately reflect current tissue activities. Conventional salivary proteomics, including the latest metaproteomic analysis by Samodova et al. [11], have highlighted overall increases in immune
proteins, mucins, and complement components in the early stages of type 2 diabetes. Although informative, such protein-level analyses represent relatively persistent aggregates of secretion, synthesis, and degradation. They rarely capture the rapid and localised biochemical events, proteolysis, epithelial desquamation, microvascular permeability, or immunological activation, that generate transient peptide fragments. By employing a rigorous peptide-focused DIA-NN approach and selectively filtering for high-confidence human peptides, our study uncovers a molecular landscape previously hidden within intact-protein datasets. Instead of broad and indistinct changes across protein families, it was observed specific and asymmetric alterations in the host salivary peptidome, indicating a deliberate reorganisation of mucosal physiology in diabetes [12]. A notable feature of the diabetic salivary peptidome is its asymmetry: a small proportion of peptides, mainly from regulatory, cytoskeletal, or epithelial-associated proteins, show reduced abundance, while over two hundred fragments display substantial increases. This pattern does not suggest a general decline in salivary gland function or a widespread increase in proteolysis; rather, it points to the targeted activation of specific biochemical processes [13,14]. Elevated peptides fall into several functionally cohesive groups: apolipoprotein fragments involved in lipid binding, complement and protease-inhibitor fragments indicating innate immune activation, mucin-derived fragments reflecting altered barrier function and viscoelasticity, and haemoglobin-derived fragments likely resulting from subclinical gingival bleeding or increased microvascular permeability. The magnitude of these increases, typically between 2- and 7-fold, demonstrates that peptide levels respond more dynamically and sensitively to metabolic and vascular stress than total protein concentrations [15]. Overall, these findings show that T2D modifies salivary composition not by universally altering secretion, but by transforming the biochemical environment of the oral cavity through coordinated changes in peptide turnover, barrier integrity, and microvascular stability.
This work differs from previous proteome-focused research through the discovery of these fragments and the mechanistic interpretation provided by structural modelling. Using AlphaFold predictions, it was showed that the diabetes-associated peptides fall into distinct structural categories that closely correspond to their biological functions. Mucin-derived peptides predominantly display disordered or loop-rich conformations, consistent with their roles in hydration, lubrication, and the formation of viscoelastic mucosal gels. Their marked enrichment in T2D saliva structurally supports the idea that diabetes alters mucosal hydration dynamics and turnover. In contrast, apolipoprotein fragments form amphipathic α-helices, the typical structural motif for lipid binding, membrane interaction, and surfactant-like properties. Their increased presence in diabetic saliva suggests not only enhanced leakage or proteolysis of plasma-derived carriers but also a shift towards a more amphipathic, lipid-interactive salivary environment that may influence microbial adherence, oral biofilm composition, or medication solubilisation [16,17]. Fragments from complement and protease- inhibitor peptides often exhibit compact interaction surfaces, indicating their potential to retain receptor binding or protease modulation capabilities even as fragments, reinforcing the idea that complement activation in T2D is not merely quantitative but functionally significant. Haemoglobin peptides retain identifiable globin helices and redox-active residues, a structural feature that aligns with their ability to influence oxidative stress or neutralise reactive species after release into the oral cavity. This structural integration enables a shift from descriptive fragment lists to a mechanistic narrative linking metabolic disorder to molecular architecture and mucosal function [18,19].
The disparity between peptide-level and intact-protein-level findings highlights a conceptual deficiency in current salivary biomarker research. Protein-level investigations, though informative, primarily identify abundant proteins whose concentrations change only after significant pathophysiological progression. In contrast, peptides are direct products of proteolytic digestion, epithelial desquamation, vascular permeability, and immunological activation, providing nearly instantaneous biochemical assessments. The peptide-level concentration of mucin fragments suggests enhanced mucosal turnover rather than simply increased mucin secretion [20,21]. The identification of certain complement-derived fragments indicates active complement cleavage rather than merely elevated baseline complement levels. The significant increase in haemoglobin peptides indicates microvascular vulnerability or gingival microbleeds that may be clinically asymptomatic yet pathophysiologically significant. Collectively, these properties suggest that the peptidome serves as a far more sensitive and mechanistically interpretable indicator of early diabetic changes than the proteome alone [22,23].
This sensitivity has distinct diagnostic implications. Numerous peptides identified in this study exhibit multi-fold differences between T2D and control saliva, a dynamic range that far exceeds conventional protein-level fluctuations in saliva or plasma. The significant scale and functional relevance of these fragments indicate that salivary peptidomics may enable non-invasive early detection or monitoring of metabolic decline. Peptide indicators may enhance or even precede glycaemic biomarkers by detecting microvascular stress, low-grade inflammation, or epithelial dysregulation before systemic glucose levels reach diagnostic thresholds. Continuous monitoring of these peptide signatures may reveal early therapeutic responses or deterioration, offering clinicians a rapid, non-invasive monitoring method that avoids the variability associated with blood sampling. Given the growing interest in saliva as a diagnostic fluid for systemic disorders, our research introduces a novel perspective by demonstrating that peptides, rather than proteins alone, may contain the most relevant diagnostic information [24,25].
This discovery also has conceptually new relevance for tissue engineering and biomaterials design. By identifying the distinct peptide motifs prevalent in diabetic saliva, hydration-enhancing mucin loops, amphipathic lipid-binding helices, immune-modulatory complement fragments, and redox-active haemoglobin segments, it provides a molecular framework for developing more authentic mucosa-mimetic bioinks. Current mucosal bioprinting techniques predominantly use generic hydrogels (e.g., gelatin, alginate, hyaluronic acid) that replicate mechanical properties but fail to emulate the biochemical complexity of native saliva or mucus [8,26]. These findings suggest that incorporating specific structural motifs from the salivary peptidome could significantly improve the physiological accuracy of oral or airway tissue models. Mucin-mimetic loops may enhance hydration and lubrication in printed tissues; amphipathic helices could influence drug solubilisation or microbial interactions; complement-like peptides might modulate immune responses; and haemoglobin-like domains could regulate redox balance in diabetic wound models. The integration of disease-specific biochemical data into biofabrication design represents an innovative interdisciplinary link between metabolically informed molecular pathology and precision- engineered mucosal structures [27,28].
This study demonstrates that examining saliva at the peptide level, rather than only at the intact protein level, uncovers a previously hidden layer of biochemical detail that is profoundly useful for understanding diabetic mucosal physiology. The integration of peptide-focused quantification, structural modelling, and translational analysis identifies distinct markers of inflammation, epithelial turnover, vascular stress, and lipid interactions that transform our understanding of salivary changes in Type 2 Diabetes. By applying these insights to diagnostics and mucosa-mimetic bioprinting, it shows how peptidomics can connect molecular pathology to practical applications, creating new opportunities for early disease detection, precision oral health monitoring, and improved design of physiologically relevant engineered tissues [29,30].
Overall, this study demonstrates that salivary peptide signatures provide a nuanced and structurally informed readout of metabolic dysregulation in type 2 diabetes. By combining high- resolution DIA-NN quantification with AlphaFold structural modelling, in advanced beyond descriptive proteomics to reveal how diabetes alters the mucosal microenvironment at the molecular level. The novelty lies not only in the specific peptides identified, but also in the dual analytical strategy employed to uncover them. This peptide-level precision revealed coordinated changes, indicative of low-grade inflammation, epithelial barrier remodelling, and systemic leakage, that were undetectable in conventional protein-level surveys. These insights have practical significance: the altered peptides identified here could serve as highly sensitive, non-invasive biomarkers for early detection and monitoring of diabetes, and the structural motifs they represent can inform the design of biomaterials (such as bioinks) that more accurately mimic the properties of healthy or diseased mucosa.
5. Conclusions
In essence, these findings provide a refined and clinically relevant perspective on the biochemical consequences of type 2 diabetes. They underscore that even in a readily accessible fluid like saliva, it is possible to detect the fingerprints of systemic disease when examined at the appropriate molecular level. This work thus expands the scope of salivary diagnostics and opens avenues for translational applications, from improved point-of-care tests to disease-informed tissue engineering. By emphasising the integration of peptidomic data with structural and functional interpretation, we aim to set the stage for future studies to exploit the rich information contained in the body’s peptide fragments. Ultimately, such approaches can deepen our understanding of metabolic diseases and enhance our ability to diagnose, monitor, and potentially therapeutically target the subtle yet significant changes occurring at our mucosal surfaces:
Supplementary Materials: The following supporting information can be downloaded at Preprints.org. Author Contributions: Conceptualization: RV, Investigation: RV. Writing-original draft: RV. Writing-review and editing.
Funding Information: This work was also financed by national funds through FCT - Fundação para a Ciência e Tecnologia, I.P., RISE (LA/P/0053/2020), for funding Institute of Biomedicine (iBiMED) (UIDB/04501/2020), and BUCCAL-PEP - 101091765.
Institutional Review Board Statement: Not applicable.
Informed Consent Statement: Not applicable.
Data Availability Statement: The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset. identifier: PXD051453.
Acknowledgments: We thank all medical professionals who are associated with our practice.
Conflicts of Interest: The author declares no conflicts of interest.
AI Disclosure Statement: Editing in English was supported by the InstaText® tool, with all suggestions carefully reviewed and approved by the authors.
References
- Dongiovanni, P.; Meroni, M.; Casati, S.; Goldoni, R.; Thomaz, D.V.; Kehr, N.S.; Galimberti, D.; Del Fabbro, M.; Tartaglia, G.M. Salivary biomarkers: novel noninvasive tools to diagnose chronic inflammation. International journal of oral science 2023, 15, 27, doi:10.1038/s41368-023-00231-6.
View at Publisher | View at Google Scholar - Abdul, N.S.; AlGhannam, S.M.; Almughaiseeb, A.A.; Bindawoad, F.A.; Alduraibi, S.M.; Shenoy, M. A review on salivary constituents and their role in diagnostics. Bioinformation 2022, 18, 1021-1028, doi:10.6026/973206300181021.
View at Publisher | View at Google Scholar - Carpenter, G.H. Salivary Factors that Maintain the Normal Oral Commensal Microflora. J Dent Res 2020, 99, 644-649, doi:10.1177/0022034520915486.
View at Publisher | View at Google Scholar - Rao, P.V.; Reddy, A.P.; Lu, X.; Dasari, S.; Krishnaprasad, A.; Biggs, E.; Roberts, C.T., Jr.; Nagalla, S.R. Proteomic Identification of Salivary Biomarkers of Type-2 Diabetes. Journal of proteome research 2009, 8, 239- 245, doi:10.1021/pr8003776.
View at Publisher | View at Google Scholar - Li, L.; Wu, J.; Lyon, C.J.; Jiang, L.; Hu, T.Y. Clinical Peptidomics: Advances in Instrumentation, Analyses, and Applications. BME frontiers 2023, 4, 0019,
View at Publisher | View at Google Scholar - Sakanaka, A.; Kuboniwa, M.; Katakami, N.; Furuno, M.; Nishizawa, H.; Omori, K.; Taya, N.; Ishikawa, A.; Mayumi, S.; Tanaka Isomura, E.; et al. Saliva and Plasma Reflect Metabolism Altered by Diabetes and Periodontitis. Front Mol Biosci 2021, Volume 8 - 2021,
View at Publisher | View at Google Scholar - Mota, C.; Camarero-Espinosa, S.; Baker, M.B.; Wieringa, P.; Moroni, L. Bioprinting: From Tissue and Organ Development to in Vitro Models. Chemical reviews 2020, 120, 10547-10607,
View at Publisher | View at Google Scholar - Käsdorf, B.T.; Weber, F.; Petrou, G.; Srivastava, V.; Crouzier, T.; Lieleg, O. Mucin-Inspired Lubrication on Hydrophobic Surfaces. Biomacromolecules 2017, 18, 2454-2462,
View at Publisher | View at Google Scholar - Chen, X.B.; Fazel Anvari-Yazdi, A.; Duan, X.; Zimmerling, A.; Gharraei, R.; Sharma, N.K.; Sweilem, S.; Ning, L. Biomaterials / bioinks and extrusion bioprinting. Bioactive Materials 2023, 28, 511-536, doi:
View at Publisher | View at Google Scholar - Fleming, J.; Magana, P.; Nair, S.; Tsenkov, M.; Bertoni, D.; Pidruchna, I.; Lima Afonso, M.Q.; Midlik, A.; Paramval, U.; Žídek, A.; et al. AlphaFold Protein Structure Database and 3D-Beacons: New Data and Capabilities. Journal of Molecular Biology 2025, 437, 168967,
View at Publisher | View at Google Scholar - Samodova, D.; Stankevic, E.; Søndergaard, M.S.; Hu, N.; Ahluwalia, T.S.; Witte, D.R.; Belstrøm, D.; Lubberding, A.F.; Jagtap, P.D.; Hansen, T.; et al. Salivary proteomics and metaproteomics identifies distinct molecular and taxonomic signatures of type-2 diabetes. Microbiome 2025, 13, 5,
View at Publisher | View at Google Scholar - Jiang, Y.; Rex, D.A.B.; Schuster, D.; Neely, B.A.; Rosano, G.L.; Volkmar, N.; Momenzadeh, A.; Peters-Clarke, T.M.; Egbert, S.B.; Kreimer, S.; et al. Comprehensive Overview of Bottom-Up Proteomics Using Mass Spectrometry. ACS measurement science au 2024, 4, 338-417,
View at Publisher | View at Google Scholar - Dallas, D.C.; Guerrero, A.; Parker, E.A.; Robinson, R.C.; Gan, J.; German, J.B.; Barile, D.; Lebrilla, C.B. Current peptidomics: applications, purification, identification, quantification, and functional analysis. Proteomics 2015, 15, 1026-1038,
View at Publisher | View at Google Scholar - Wang, L.; Main, K.; Wang, H.; Julien, O.; Dufour, A. Biochemical Tools for Tracking Proteolysis. Journal of proteome research 2021, 20, 5264-5279,
View at Publisher | View at Google Scholar - Masaki, T.; Kodera, Y.; Terasaki, M.; Fujimoto, K.; Hirano, T.; Shichiri, M. GIP_HUMAN [22–51] is a new proatherogenic peptide identified by native plasma peptidomics. Scientific reports 2021, 11, 14470,
View at Publisher | View at Google Scholar - Zhu, H.L.; Atkinson, D. Conformation and Lipid Binding of a C-Terminal (198−243) Peptide of Human Apolipoprotein A-I. Biochemistry 2007, 46, 1624-1634,
View at Publisher | View at Google Scholar - Wang, L.; Atkinson, D.; Small, D.M. Interfacial Properties of an Amphipathic α-Helix Consensus Peptide of Exchangeable Apolipoproteins at Air/Water and Oil/Water Interfaces*. Journal of Biological Chemistry 2003, 278, 37480-37491,
View at Publisher | View at Google Scholar - Hamers, S.; Abendstein, L.; Boyle, A.L.; Jongkees, S.A.K.; Sharp, T.H. Selection and characterization of a peptide-based complement modulator targeting C1 of the innate immune system. RSC Chem Biol 2024, 5, 787-799,
View at Publisher | View at Google Scholar - Berg, T. Modulation of Protein–Protein Interactions with Small Organic Molecules. Angewandte Chemie International Edition 2003, 42, 2462-2481,
View at Publisher | View at Google Scholar - Niebla-Cárdenas, A.; Bueno-Hernández, N.; Hernández, A.-P.; Fuentes, M.; Méndez-Sánchez, R.; Arroyo- Anlló, E.M.; Orera, I.; Lattanzio, G.; Juanes-Velasco, P.; Arias-Hidalgo, C.; et al. Potential protein biomarkers in saliva for detection of frailty syndrome by targeted proteomics. Mechanisms of Ageing and Development 2024, 221, 111974,
View at Publisher | View at Google Scholar - Chen, Y.; Qi, Y.; Lu, W. Endogenous Vasoactive Peptides and Vascular Aging-Related Diseases. Oxid Med Cell Longev 2022, 2022, 1534470,
View at Publisher | View at Google Scholar - Dunkelberger, J.R.; Song, W.-C. Complement and its role in innate and adaptive immune responses. Cell research 2010, 20, 34-50,
View at Publisher | View at Google Scholar - Kidmose, R.T.; Laursen, N.S.; Dobó, J.; Kjaer, T.R.; Sirotkina, S.; Yatime, L.; Sottrup-Jensen, L.; Thiel, S.; Gál, P.; Andersen, G.R. Structural basis for activation of the complement system by component C4 cleavage. Proceedings of the National Academy of Sciences 2012, 109, 15425-15430,
View at Publisher | View at Google Scholar - Hu, D.; Wu, Y.; He, D.; Rong, J.; Sun, M.; Yu, Z.; Wu, L. Salivary biomarkers in diabetes mellitus diagnostics: A bibliometric analysis. Medicine (Baltimore) 2025, 104, e43760,
View at Publisher | View at Google Scholar - Wazwaz, F.; Saloom, H.; Houghton, J.W.; Cobourne, M.T.; Carpenter, G.H. Salivary peptidome analysis and protease prediction during orthodontic treatment with fixed appliances. Scientific reports 2023, 13, 677,
View at Publisher | View at Google Scholar - Bej, R.; Haag, R. Mucus-Inspired Dynamic Hydrogels: Synthesis and Future Perspectives. Journal of the American Chemical Society 2022, 144, 20137-20152,
View at Publisher | View at Google Scholar - Hernandez, A.; Hartgerink, J.D.; Young, S. Self-assembling peptides as immunomodulatory biomaterials. Front Bioeng Biotechnol 2023, 11, 1139782,
View at Publisher | View at Google Scholar - Zhang, S.-K.; Song, J.-w.; Gong, F.; Li, S.-B.; Chang, H.-Y.; Xie, H.-M.; Gao, H.-W.; Tan, Y.-X.; Ji, S.-P. Design
View at Publisher | View at Google Scholar - of an α-helical antimicrobial peptide with improved cell-selective and potent anti-biofilm activity. Scientific reports 2016, 6, 27394,
View at Publisher | View at Google Scholar - Desai, P.; Donovan, L.; Janowid, E.; Kim, J.Y. The Clinical Utility of Salivary Biomarkers in the Identification of Type 2 Diabetes Risk and Metabolic Syndrome. Diabetes Metab Syndr Obes 2020, 13, 3587-3599,
View at Publisher | View at Google Scholar - Yeruva, T.; Yang, S.; Doski, S.; Duncan, G.A. Hydrogels for Mucosal Drug Delivery. ACS Appl Bio Mater 2023, 6, 1684-1700,
View at Publisher | View at Google Scholar

 Clinic
Clinic
