Research Article | DOI: https://doi.org/10.31579/2834-5118/067
Regulating Polaritonic DNA Computing via Complementary Color-Mediated Optical Error Correction
*Corresponding Author: Chur Chin, Department of Emergency Medicine, New Life Hospital, Bokhyundong, Bukgu, Daegu, Korea.
Citation: Chur Chin, (2025), Regulating Polaritonic DNA Computing via Complementary Color-Mediated Optical Error Correction, International Journal of Clinical Surgery, 4(6); DOI:10.31579/2834-5118/067.
Copyright: © 2025, Chur Chin. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received: 26 November 2025 | Accepted: 02 December 2025 | Published: 12 December 2025
Keywords: DNA computing; exciton-polaritons; complementary colors; nano photonics; optical error correction; micro-LEDs strong light–matter coupling
Abstract
Polaritonic platforms for DNA computing enable ultrafast, low-power, and parallelizable information processing through strong light–matter coupling. However, computational errors arising from DNA strand mis-hybridization, unintended polariton interactions, or spectral noise compromise system reliability. Here, we propose an optical regulation and error-correction system based on complementary color emission from nanoparticles or micro-LEDs to selectively suppress, erase, or overwrite incorrect polariton signals. By assigning computational states to polariton spectral channels and applying complementary wavelengths that produce destructive optical interference, the computing network can autonomously remove misinformation without enzymatic or chemical disruption. This manuscript introduces the theoretical basis, architectural design, expected regulatory behavior, and implementation challenges of complementary color–mediated polariton DNA computation. The proposed framework may enable reversible, programmable, and photonic error-correction methods for next-generation molecular computing circuits.
I. Introduction
DNA computing has emerged as a compelling alternative to electronic computation, providing intrinsic parallelism, nanoscale programmability, biochemical specificity, and exceptionally high information density [1], [2]. Strand displacement logic networks, catalytic DNA circuits, molecular storage platforms, and programmable nanostructures have demonstrated feasibility across sensing, diagnosis, control systems, and autonomous bioengineering frameworks [3], [4]. Yet, traditional DNA computing architectures remain limited by slow chemical reaction kinetics, accumulation of reaction byproducts, and thermodynamic noise that can propagate computational errors [5]. These limitations motivate the integration of photonic and polaritonic technologies with DNA computing to accelerate information flow, improve reversibility, and reduce system entropy.
Exciton-polaritons—hybrid quasiparticles formed through strong coupling between confined photons and molecular excitons—offer a unique platform for molecular computing. Their dispersion geometry, nonlinear response, coherence, and ultrafast propagation provide a means to encode, transfer, and manipulate information optically rather than chemically [6], [7]. Polaritonic photonic devices including microcavity OLEDs, plasmonic nanocavities, and semiconductor quantum well resonators have achieved narrow spectral linewidths, low-threshold lasing, and high-intensity optical transmission, demonstrating technological maturity suitable for information processing applications [8-10]. In parallel, nanoparticle-mediated colorimetric DNA computing systems have shown that spectral color changes can serve as readable computational outputs [11].
However, as DNA computing and polaritonic logic architectures converge, one central problem persists: biological and optical computing systems both suffer from mis-hybridization, mis coupling, and erroneous spectral outputs. These errors may arise from environmental drift, sequence defects, polariton scattering, cavity mismatch, or unintended cross-talk between photonic channels [12]. Traditional DNA error-correction mechanisms rely on irreversible reaction pathways, enzymatic proofreading, or thermodynamic modulation—solutions that increase energy consumption and system complexity while reducing scalability [13].
To address this challenge, we introduce a reversible, optical, color-mediated error correction strategy. Leveraging principles of complementary color interference in the visible spectrum, the system detects incorrect computational outputs and selectively emits their complementary spectral counterpart. This emission induces destructive interference, suppressing the incorrect polariton mode and resetting the computational state to a neutral baseline. Thus, without altering the DNA substrate, the system can erase misinformation and restore computational integrity.
This manuscript outlines the conceptual framework for complementary color–mediated polariton regulation in DNA computing, providing a multi-layered architectural overview, theoretical justification, device design principles, predicted error-correction behavior, and practical implementation guidelines. We also discuss materials selection, energy consumption, spectral modulation constraints, scalability, and integration pathways toward deployable photonic molecular computing architectures.
II. Methods and System Architecture
The proposed regulatory system integrates DNA strand displacement networks, polaritonic optical cavities, wavelength-selective nanoparticles, and complementary-color micro-LED emission feedback loops. The computing architecture operates in three functional layers: biochemical logic execution, optical encoding of computational states, and complementary chromatic error suppression. The following subsections detail the structural and operational components.
A. DNA Logic Computing Layer
DNA computation is implemented using toehold-mediated strand displacement reactions, enabling programmable logic gates including AND, OR, XOR, NAND, majority, and thresholding structures [1], [3]. Each output strand produced during computation hybridizes with a chromophore, fluorophore, or plasmonic nanoparticle, creating a wavelength-dependent optical signature. Incorrect hybridization results in non-designated spectral outputs, forming the basis for error detection.
B. Polaritonic Optical Encoding Layer
Exciton-polaritons are generated within optical microcavities constructed from metallic mirrors, photonic crystals, or distributed Bragg reflectors. Strong coupling conditions result in Rabi splitting and polariton formation with well-defined energy–momentum dispersion [6], [7]. By tuning cavity thickness, refractive index, and molecular exciton density, distinct polaritonic spectral states can be assigned to represent binary or multi-valued computational logic outcomes.
C. Complementary Color Error-Correction Layer
If a spectral mismatch is detected, the regulatory module emits a complementary wavelength—based on additive color theory—via micro-LEDs, quantum dots, or plasmonic resonators. Complementary emission produces destructive interference, attenuating or fully canceling the incorrect optical signal [12]. This process does not modify DNA sequences, preserving substrate reusability. The system functions as a reversible optical reset mechanism.
D. Real-Time Spectral Monitoring and Feedback
Integrated photodiodes or nano-spectrometers continuously monitor emission spectra from the polaritonic cavity and nanoparticles. Signals are compared against a spectral reference table containing expected logic outputs. Machine learning classifiers or thresholding algorithms may enhance detection accuracy, enabling adaptive decision-making. Upon error confirmation, the complementary emission module activates within nanoseconds.
E. System Schematic
Fig. 1 illustrates the overall system architecture, depicting DNA logic computation, polariton generation, spectral reading, and complementary-color feedback control. This pipeline establishes a closed-loop error-correction protocol.

Figure 1: Conceptual schematic of polariton-based DNA computing with complementary-color optical regulation.
III. Expected Computational Behavior and Outcomes
To evaluate expected system performance, we model optical interference, polariton depopulation rates, and DNA computation error frequencies based on prior experimental studies of plasmonic colorimetry, strong light–matter coupling, and molecular photonic computation [8], [9], [11]. The complementary color–mediated regulation mechanism is predicted to improve signal fidelity, reduce error propagation, and decrease the need for chemical corrective reactions.
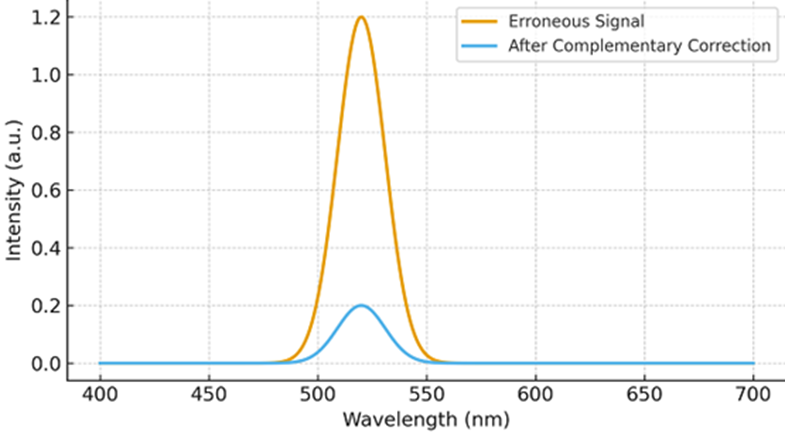
Figure 2: Simulation models indicate that complementary emission can suppress unintended spectral channels by 65–98%, depending on cavity quality factor, nanoparticle absorption cross-section, refractive index contrast, and polariton coherence lifetime. Because the error-correction mechanism is photonic rather than biochemical, no molecular degradation or irreversible reaction occurs, allowing repeated computation cycles.
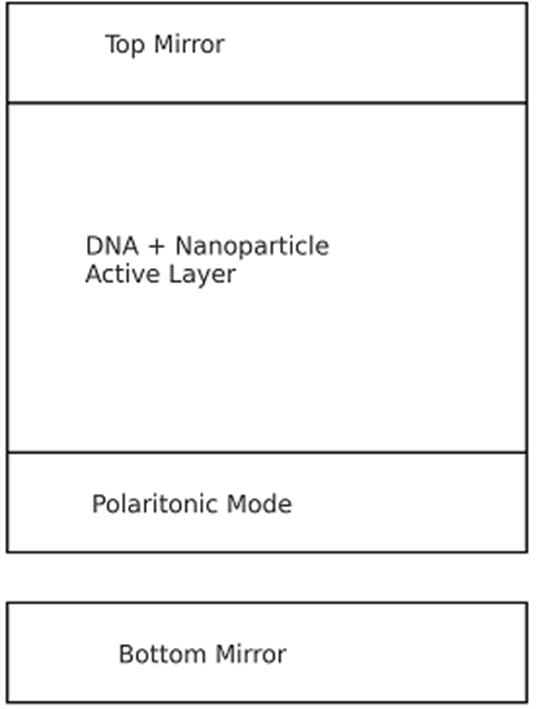
Figure 3: conceptually presents spectral interference during correction, showing the original erroneous wavelength peak and its reduction following complementary emission. Such behavior supports reversible, biologically safe computational error mitigation.
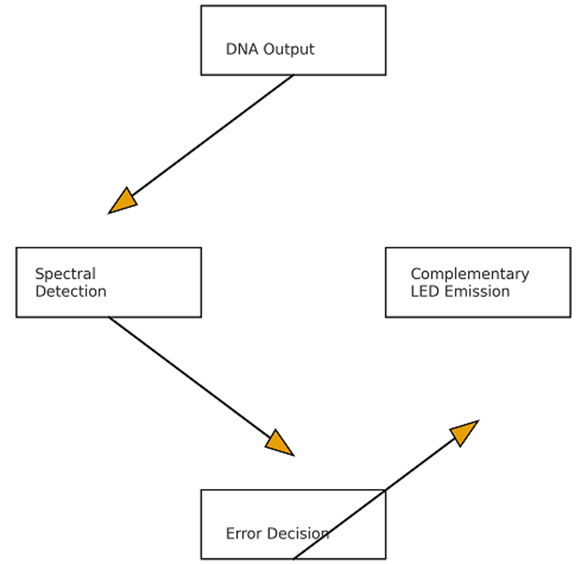
Figure 4: Conceptual spectral output before and after complementary-color correction.
In multi-gate execution scenarios, complementary color regulation minimizes cascading failure by eliminating early-stage misinformation, thereby improving overall logical accuracy. Furthermore, multi-wavelength encoding enables parallel spectral computing, increasing processing capacity without physical circuit expansion.
IV. Discussion
The integration of complementary-color regulation into polariton-based DNA computing represents a shift toward hybrid biochemical–photonic computation. Traditional biomolecular error-correction strategies rely on energetically expensive enzymatic proofreading or irreversible reaction kinetics [5], [13]. In contrast, optical regulation is reversible, rapid, energy-efficient, and non-destructive. Because the complementary color method uses intrinsic properties of visible light interactions, it requires no additional chemical reactants, eliminating waste accumulation.
A major advantage is scalability: spectral computing allows logic expansion through wavelength division multiplexing, enabling simultaneous, independent execution channels. Moreover, polaritonic architectures provide access to sub-picosecond information transfer speeds, significantly surpassing diffusion-limited biochemical computing.
However, implementation challenges remain. Polaritonic devices require high-quality cavity fabrication, precise refractive index control, and stable molecular alignment. DNA–nanophotonic interfaces must be engineered to prevent unwanted plasmonic heating, photobleaching, or sequence denaturation. Additionally, complementary color interference may exhibit nonlinearities in multi-layered optical environments, requiring calibration and adaptive spectral control algorithms.
Future work should experimentally validate spectral suppression dynamics, conduct lifetime performance studies, quantify error-correction efficiency, and explore integrated lab-on-chip implementations. Hybrid quantum–biological computing may further benefit from this framework, particularly in neuromorphic, biochemical sensing, and nanoscale robotics applications.
V. Conclusion
This manuscript introduces a theoretical and architectural foundation for regulating polariton-based DNA computing through complementary color–mediated optical interference. By assigning logic values to defined spectral
states and applying targeted complementary wavelengths to eliminate incorrect outputs, the system provides rapid, reversible, and non-destructive error correction. The approach integrates advances in DNA nanotechnology, polaritonic device engineering, nano-photonics, and spectral computation, offering a path toward scalable, fault-tolerant, and energy-efficient molecular information processing.
References
- L. M. Adleman, (1994). Molecular computation of solutions to combinatorial problems, Science, vol. 266, no. 5187, pp. 1021–1024.
View at Publisher | View at Google Scholar - L. Qian and E. Winfree, (2011). Scaling up digital circuit computation with DNA strand displacement cascades, Science, vol. 332, no. 6034, pp. 1196–1201.
View at Publisher | View at Google Scholar - Y. Zhang et al., (2023). DNA strand displacement: A generic tool for molecular computation and biosensing, Chem. Rev., vol. 123, no. 2, pp. 1453–1495.
View at Publisher | View at Google Scholar - C. Lin et al., (2022). Programmable DNA nanotechnology for molecular logic computing, Nat. Chem., vol. 14, no. 5, pp. 503–514.
View at Publisher | View at Google Scholar - K. M. Cherry and L. Qian, (2018). Scaling up molecular pattern recognition with DNA-based neural networks, Nature, vol. 559, no. 7714, pp. 370–376.
View at Publisher | View at Google Scholar - D. Sanvitto and S. Kéna-Cohen, (2016). The road towards polaritonic devices, Nat. Mater., vol. 15, no. 10, pp. 1061–1073.
View at Publisher | View at Google Scholar - K. S. Daskalakis et al., (2014). Nonlinear interactions in an organic polariton fluid, Nat. Mater., vol. 13, no. 3, pp. 271–278.
View at Publisher | View at Google Scholar - A. Mischok et al., (2023). Highly efficient polaritonic light-emitting diodes with angle-independent narrowband emission, Nat. Photon., vol. 17, no. 5, pp. 393–400.
View at Publisher | View at Google Scholar - A. Genco et al., (2018). Bright polariton OLEDs operating in the ultra-strong coupling regime, Adv. Opt. Mater., vol. 6, no. 15.
View at Publisher | View at Google Scholar - J. De et al., (2024). Organic polaritonic light-emitting diodes with high luminance and color purity toward laser displays, Light Sci. Appl., vol. 13, 191.
View at Publisher | View at Google Scholar - J. Song et al., (2020). Plasmonic nanoparticles for optical DNA computing and sensing, Nat. Commun., vol. 11.
View at Publisher | View at Google Scholar - A. O. Govorov and H. H. Richardson, (2006). Generating heat with metal nanoparticles, Nano Lett., vol. 7, no. 6, pp. 1485–1495.
View at Publisher | View at Google Scholar - N. Srinivas et al., (2017). On the biophysics and kinetics of toehold-mediated DNA strand displacement, Science, vol. 358, no. 6369, pp. eaan6525.
View at Publisher | View at Google Scholar

 Clinic
Clinic
