Research Article | DOI: https://doi.org/10.31579/2834-8664/026
First Isolate of Orientia Tsutsugamushi from Vellore, South India
- Janaki Kumaraswamy 1
- Agilandeeswari Kirubanandan 1
- Lakshmi Surya Nagarajan 1
- Karthik Gunasekaran 2
- Abhilash KPP 3
- John Antony Jude Prakash 1*
¹ Department of Clinical Microbiology, Christian Medical College, Vellore, Tamil Nadu, India.
² Department of Medicine, Christian Medical College, Vellore, Tamil Nadu, India.
³ Emergency Medicine, Christian Medical College, Vellore, Tamil Nadu, India.
*Corresponding Author: John Antony Jude Prakash, Department of Clinical Microbiology, Christian Medical College, Vellore, Tamil Nadu, India.
Citation: Janaki Kumaraswamy, Agilandeeswari Kirubanandan, Lakshmi S. Nagarajan, Karthik Gunasekaran, Abhilash KPP, et al, (2023), First isolate of Orientia tsutsugamushi from Vellore, South India, International Journal of clinical and Medical Case Reports, 2(3); Doi:10.31579/2834-8664/026
Copyright: © 2023, John Antony Jude Prakash. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Received: 05 June 2023 | Accepted: 23 June 2023 | Published: 30 June 2023
Keywords: speciation-extinction analyses; biogeography; climate; animals and plants; regional biota; species diversity and richness
Abstract
Background: Scrub typhus a common cause of acute febrile illness in India caused by Orientia tsutsugamushi an obligate intracellular bacterium requiring cell culture for isolation. Cell lines like Vero and L929 are most suitable for isolating and maintaining this organism. This study was undertaken to isolate and characterize of Orientia tsutsugamushi from whole blood samples at a tertiary care centre in Southern India.
Methods: The PBMCs (peripheral blood mononuclear cells) collected from scrub typhus positive (47kDa qPCR positive) patients were inoculated into Vero and L929 cell line at 80% confluence for primary isolation. The inoculated flasks were incubated at 37⁰C with 5% CO2 for 30 days and examined for presence of Orientia tsutsugamushi on the day 10, 15, 20 post- inoculation and everyday thereafter for a maximum of 30 days post inoculation. The scrapings were subjected to Giemsa staining, IFA, 47kDa qPCR and transmission electron microscopy (TEM). The isolates were passaged 3-4 times to ensure viability and then stored in DMEM with 10% FBS (-80℃). Genotyping of the isolates was performed by amplifying a 650 bp segment of the TSA 56 (type specific antigen 56) gene.
Results: Amongst the 50 samples inoculated, three were culture positive as confirmed by 47 kDa qPCR at 24th day of inoculation. This was further confirmed by Giemsa, IFA staining and TEM. The 650bp amplicons showed 99.5 to 100% homology with Orientia tsutsugamushi MW604716, MH003839, MW604718, MW604717, MH922787 and
MH003838 strains. Phylogenetic analysis revealed that 2 isolates belong to TA763 genotype and one belongs to Gilliam genotype.
Conclusions: We have successfully isolated and characterised the Orientia tsutsugamushi for the first time at our centre from PBMCs. Based on the partial TSA56 gene sequence our isolates belongs to TA763 and Gilliam genotype. More number of samples are being processed for identifying further isolates followed by genomic analysis.
Introduction
Orientia tsutsugamushi is an obligate intracellular bacteria, the etiological agent of vector borne zoonotic disease Scrub typhus [1]. Orientia tsutsugamushi strains are classified into several subgroups based on the antigenic variations of the major surface protein called type specificantigen 56 (TSA 56) [2,3].
The infection is transmitted to humans (accidental dead-end host) by the bite of an infected larval stage of trombiculid mites commonly called as chiggers [1]. In humans, O. tsutsugamushi invades the macrophages and vascular endothelial cells, escapes from the phagosomes, and propagates in the cytosol, inducing an acute febrile state. It often causes a deadly illness, if not treated with appropriate antibiotics, but the molecular pathogenicity is still poorly understood and no vaccine is currently available [4,5].
The scrub typhus diagnosis are mostly based on serological techniques. Currently recognized serological reference standard test is the indirect immunofluorescence assay (IFA) [6]. The scrub typhus IgM ELISA also has a similar sensitivity and specificity to the IFA. This assay utilizes the O. tsutsugamushi-specific recombinant 56-kDa antigen [7,8]. PCR, especially real-time PCR (qPCR) is most useful for diagnosis in the early stage of scrub typhus infection. The common targets of these assays are the genes encoding the 56-kDa antigen and 47-kDa surface antigen,16S rRNA and groEL genes [9-12].
Isolation of O. tsutsugamushi in cell culture, though technically demanding, is useful for understanding the biology includingstudying the genomicsand determining antimicrobial susceptibility [13]. Vero andL929 cell lines have been traditionally used to isolateand propagate O. tsutsugamushi [15, 16]. Growth of the organism is slow (20- 30 days), therefore the results are of little use to the patient as well as the treating clinician [14,15]. However, it is important to obtain isolates of this bacteria to study the biology of the organisms including pathogenesis, virulence factors, host cell interactions, and genomic features. Further the availability of isolates will be the source of nucleic acids which can be used as positive control for various PCR assays, whole genome sequencing [15-17]. The culture isolates can also be useful to develop newer diagnostics and vaccines, which are region specific[15].
Only few studies are available globally on isolation of Orientia tsutsugamushi and none from India (Prakash JA personal communication). We, therefore proceeded to establish a standard protocol for isolation, characterisation and maintenance of Orientia tsutsugamushi fromwhole blood samples at our centre.
Methodology
Sample collection and processing
Whole blood (8 ml) was collected in cell processing tubes (CPT) with sodium heparin (BDVacutainer® CPT™, Franklin Lakes, NJ, USA) from clinically suspected scrub typhus patients. The collected samples were centrifuged at 1600 RCF for 20 minutes at room temperature. The separated plasma was aliquoted and stored; the monocyte layer was pipetted carefully and aliquoted into two tubes with the addition of equal volume of MEM/DMEM. One aliquot of monocytes was immediately transported to cell culture laboratory and inoculated into cell culture; other was used for DNA extraction using the Promega Wizard® Genomic DNA Purification kit (Promega Corporation, Madison,WI, USA) as per the manufacturer’s protocol. The extracted DNA was quantified using Nanodrop Spectrophotometer (ThermoFisher Scientific, Waltham, MA, USA) and stored at -80ºC for molecular assays.
Isolationof Orientia tsutsugamushi in cell culture:
The Vero and L929 cell line were maintained according to the protocol described by National Centre for Cell Science (NCCS), Pune. The cells were grown in 25 cm2 flasks (Greiner) with growth medium [MEM (minimalessential medium), Himedia, Thane, Maharastra, India] with 10% foetal bovine serum (FBS, Merck, Rahway, NJ, USA) at 37℃ and in a humidified atmosphere containing 5% CO2. At 80-100% confluence the cells were sub-cultured into a new flask (1:5) and maintained.
The PBMCs with MEM were directly inoculated into a 10 cm2 Vero and L929 cell line at 80% confluence. The inoculated flasks were kept in a shaker for 30 minutes, then at 37℃ for 2 hours to facilitate infection. The inoculum was removed and replaced with fresh MEM with 2
Results
Of the 50 samples inoculated into cell culture, Orientia tsutsugamushi was isolated in Vero cells from 3 (confirmed by 47 kDa qPCR) after 24th day of inoculation (refer Table 1). The positive samples (Vellore isolate 1-3) were sub-cultured into 25cm2 flask. The three isolates were genotyped by using the 56 kDa type specific antigen partial gene sequence (650 bp). BLAST analysis of this partial sequence showed 99.5 to 100% homology with MW604716, MH003839, MW604718, MW604717, MH922787 and MH003838 Orientia tsutsugamushi strains. Phylogenetic analysis revealed that the Vellore isolates 1 and 2 belong to TA763 whereas the Vellore isolate 3 belongs Gilliam genotype (Figure 4).
Among the three, 2 isolates were cryopreserved and one isolate was maintained for further morphological analysis such as Giemsa, IFA and TEM (Transmission Electron Microscopy).
Giemsa staining revealed purple colour small rod-shaped organisms in Vero cells Figure 2. IFA showed bright green stained organisms as shown in Figure 3. Further, we analysed the ultrastructural morphology of Orientia tsutsugamushi in infected Vero cell by TEM. TEM showed Orientia tsutsugamushi in infected Vero cell cytoplasm, the bacterium was round to oval shape with characteristic double membrane (Figure 4B and 4E: indicated by black star), whereas the mitochondria are smaller, darker, spherical/elliptical with cristae seen in higher magnification (Figure 4C and 4D: white arrow head). Extracellular Orientia tsutsugamushi are shown in Figure 4F (black arrows).
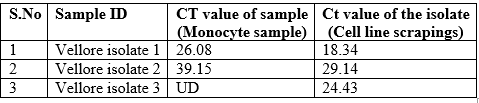
Table 1. Cultureisolates positive for 47kDa q PCR
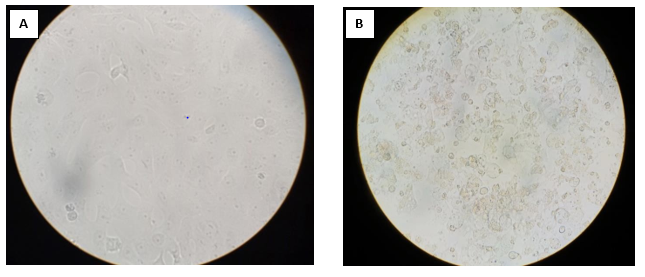
Figure 1. A. Normal Vero cell line, B. Cytopathic effect of Vellore isolate 1 in Vero cell line (Magnification 400X)
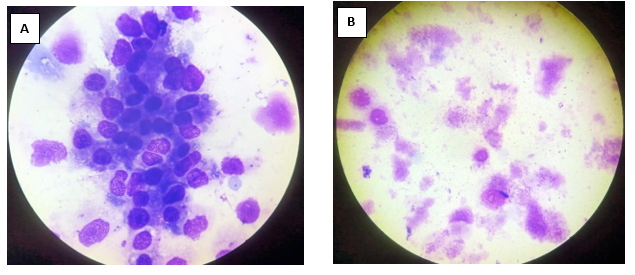
Figure 2: Giemsa staining of Vellore isolate 1 isolate. A. Negative control (uninfected Vero cells) B. Orientiatsutsugamushi infected Vero cells
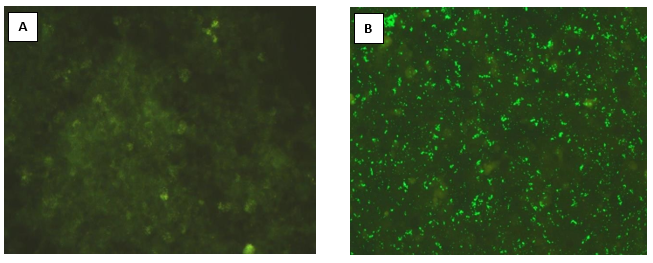
Figure 3: Indirect Fluorescence Assay of Vellore isolate 1 isolate: A. Negative control (unifected Vero cells), B. Orientia tsutsugamushi infected Vero cells
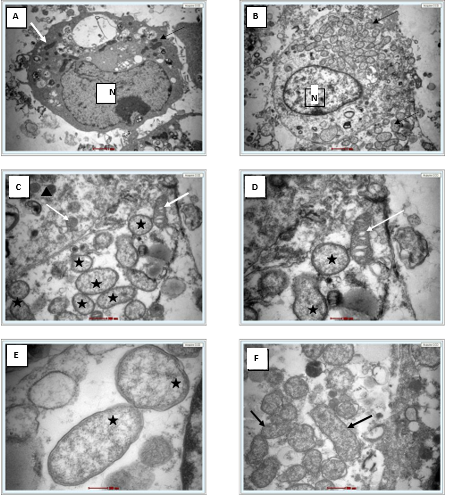
Figure 4. A. Single uninfected Vero cell (6000X) white arrow indicates mitochondria, N- Nucleus. B. Single infected Vero cell (6000X) showed intact cytosolic Orientia tsutsugamushi (arrows). C. At 16500X, Intact bacterium (black star) and mitochondria (white arrow), D. At 26500X, bacterium (black star) and mitochondria with cristae (white arrow), E. At 43000X, Orientia tsutsugamushi featuring a double membrane and diffuse cytoplasm (black star). F. Extracellular free Orientia tsutsugamushi (black arrow) 16500X.
Phylogenetic tree:
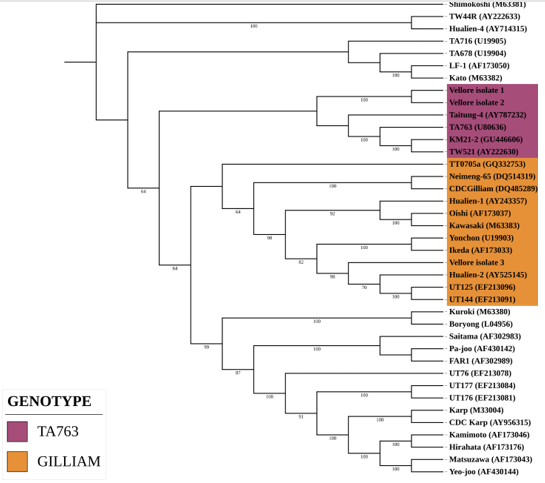
Figure 5. Phylogenetic analysis of Vellore isolates
Discussion
The in vitro isolation of Orientia tsutsugamushi from clinical samples is important to understand the genomic diversity of the pathogen for development of vaccines and diagnostics. In this study, we have successfully isolated the Orientia tsutsugamushi from whole blood sample. The use of 10cm2 flask for initial isolation is a simplified method to get an isolate from minimal amount of sample. The 10 cm2 culture tube has a wide mouth opening for easy manipulation to sample the infected cell line /supernatant.
Initially, MEM was used for isolation but MEM depleted faster, this required frequent media change. Hence, we switched to DMEM which is more suitable for isolation and maintenance of Orientia tsutsugamushi as it has longer incubation period. DMEM is complex media ontains four fold increased concentrations of amino acids and vitamins. Cells grow faster and deplete the nutrients slower in DMEM.
Phylogenetic analysis of the partial gene sequence (Horinouchi protocol) which covers the variable domain I, II and III revealed our isolates belongs to TA763 and Gilliam genotype (10,18).
One isolate was morphologically characterised by Giemsa staining, IFA and Electron microscopy. The size and shape of the bacterium resembles mitochondria, the cristae and the difference in electron density helped to identify the Orientia tsutsugamushi (Figure 4C and D) as described by Paris and co-workers and cited in the literature by Gemma Vincent (17).
Conclusion
We have successfully isolated and characterised Orientia tsutsugamushi for the first time at our centre and most likely in India from PBMCs. Based on the partial TSA56 gene sequence our isolates belongs to TA763 and Gilliam genotype. Complete 56kDa and whole genome analysis is needed to definitively identify the genotype. More number of samples from suspected cases are being processed for isolation in cell culture to obtain information regarding the genotypes of Orientia tsutsugamushi circulating in and around Vellore.
Author contributions: Conceptualization and methodology: J.K., K.G., A.K.P.P., J.A.J.P.; Investigations and Data acquisition: J.K., A.K., L.S.N.; Analysis and data interpretation: J.K., A.K., L.S.N., J.A.J.P., Drafting the manuscript: J.K., L.S.N.; Critical revision of the manuscript: K.G., A.K.P.P., J.A.J.P.; Supervision and funding acquisition: J.A.J.P.
Conflicts of Interest: The authors declare that they have no conflicts of interest.
Funding: The study was funded by Indian Council of Medical Research (ICMR) grant awarded to Prakash JAJ (Grant no.: ZON/42/2019/ECD-II).
Ethics approval: The study was approved by Institution Review Board (Silver, Research and Ethics Committee) of the Christian Medical College (CMC), Vellore, India. IRB Min. No. 11942, Dated 27.03.2019
Acknowledgements: None
References
- Watt G, Parola P. Scrub typhus and tropical rickettsioses. Curr Opin Infect Dis. 2003 Oct;16(5):429–36.
View at Publisher | View at Google Scholar

 Clinic
Clinic
