Research Article | DOI: https://doi.org/10.31579/2834-8486/013
DRUG REPOSITIONING, AN APPROACH FOR IDENTIFICATION OF NEW THERAPEUTICS
- Shaheen Yaseen
- Saima Akram *
Institute Of Biochemistry and Biotechnology, University of The Punjab, Lahore, Pakistan.
*Corresponding Author: Saima Akram, Institute of Biochemistry and Biotechnology, University of The Punjab, Lahore, Pakistan.
Citation: Shaheen Yaseen, Saima Akram. (2023), DRUG REPOSITIONING, AN APPROACH FOR IDENTIFICATION OF NEW THERAPEUTICS. Biomedical and Clinical Research, 2(6); DOI:10.31579/2834-8486/013
Copyright: © 2023, Saima Akram. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Received: 09 November 2023 | Accepted: 29 November 2023 | Published: 21 December 2023
Keywords: drug repositioning; drug repurposing; drug re profiling; de novo drug discovery; computational approach; experimental approach; synergistic drug repositioning
Abstract
Drug repositioning of FDA approved drugs is an innovation stream of drug discovery. This review elaborates the advantages, approaches and challenges of drug repositioning. This low cost, less risky and less time-consuming drug repositioning includes effective computational and experimental approaches to identify new indications for existing drugs. Similar to drug discovery, there are various challenges to repurpose a drug such as selection of an appropriate approach and target population, intellectual property (IP) protection and etc. This review also focuses on history and success stories of drug repositioning. An alternative approach, drug combination of two or multiple drugs has dramatically increased the success rate of drug repositioning. This synergistic drug repositioning is a promising strategy to combat various diseases such as infectious diseases, cancer, neurological disorders and many rare diseases. Various examples of successful synergistic drug repositioning have been described in this review. Finally, we can say that synergistic drug repositioning provides a new way to drug discovery research.
Introduction
In the 1870s, Paul Ehrlich introduced the idea of “chemoreceptor”. Since that time, scientists are searching for those compounds that interact with specific targets to treat various diseases. These medications proved beneficial but this one-drug-one-target model has some limitations (Kola and Landis 2004). Now, our current challenge is to find “master keys” that can interact with multiple targets (Medina Franco, Giulianotti et al. 2013). One strategy to win the challenge is drug repositioning (also known as drug repurposing and drug reprofiling). Drug repositioning is defined as identifying and developing new uses for existing drugs (Ashburn and Thor 2004). It also involves exploitation of established drugs that have already been approved for treatment. 1.1 Stages Of De Novo Drug Discovery And Development And Drug Repositioning Denovo drug discovery includes six stages that are target discovery, discovery & screening, lead optimization, ADMET (absorption, distribution, metabolism, excretion and toxicity), development and registration. While repositioning process of a drug includes four stages are compound identification, compound acquisition, development and registration as illustrated in figure 3 (Ashburn and Thor 2004).
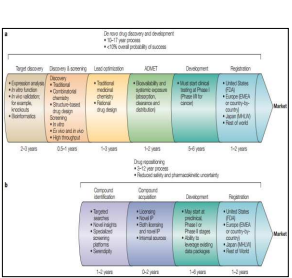
Figure 3: Stages of de novo drug discovery and development and drug repositioning.
de novo drug discovery and development b) drug repositioning. EMEA, European Medicines Agency; FDA, Food and Drug Administration; IP, intellectual property; MHLW, Ministry of Health, Labour and Welfare (Ashburn and Thor 2004) . 1.2 Advantages Of Drug Repositioning Drug repositioning is an effective strategy. Development of a new drug requires time, cost, effort and it is a highly risky process (Xue, Li et al., 2018). Drug repositioning has following advantages · Drug repositioning requires less time for development as compared to development of a new drug. According to a report of Eastern Research Group (ERG), development process of a new drug requires 10-15 years. On average, 1-2years are required for the identifications of new targets and 8 years are required for the development of repositioned drug (Sertkaya et al., 2014; Xue, Li et al., 2018). · According to a report of Food and Drug Administration (FDA), expenditure on the development of new drugs is increasing continuously while the number of approved drugs by FDA is decreasing as depicted in figure 1 (Ashburn and Thor 2004). Expenditure on conventional drug development method is high as compared to that on drug repositioning. The cost for the development of new drug by conventional methods is $12billion and that for repositioning is
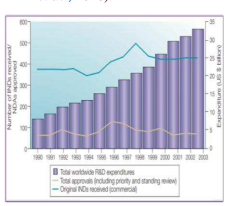
Figure 1: The growing productivity gap in the biopharmaceutical industry (Ashburn and Thor 2004).
Reduction in timelines and cost for pharmaceutical research results in the increase in risk. Drug repositioning has reduced this risk because repurposed drugs have passed all phases of clinical development and their safety profiles are known. A reward-risk diagram is often used to compare repositioning and conventional process of drug development (Ashburn and Thor 2004). Repositioned drugs have high reward and low risk as shown in figure 2 (Xue, Li et al., 2018).
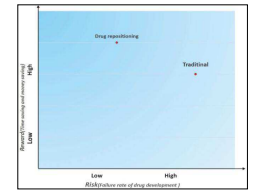
Figure 2: The risk-versus-reward trade-off between different drug development strategies (Xue, Li et al., 2018). · Drug repositioning is less laborious. Much effort is required for the development of new drug by conventional method. Success rate of the development of new drug is only 2.1% (Yeu et al.,2015; Xue, Li et al., 2018). 1.3 Approaches Of Drug Repositioning Historically many repositioned drug candidates have been discovered serendipitously. There are various approaches of drug repositioning as depicted in figure 4 (Grinnan, Trankle et al. 2019). Computational approach includes Drug combination prediction, EHR based drug repositioning, Fragment-based drug repositioning, Genome-based drug repositioning, Network-based drug repositioning, Neural network-based drug repositioning, Off-targeting data driven repositioning, Pathway based repositioning, Protein-protein interaction driven prioritization, Protein-small molecule interactions, Structure based drug repositioning, Systematic drug repositioning, Text-mining driven drug repositioning and Topic modeling. Experimental approach includes Chemical genomics, Chemical systems biology, Clinical side-effects, Genome-wide association studies (GWAS), Medical genetics, MicroRNA (miRNA) expression, Network medicine, Serological marker, Small interference RNA (SiRNA) screening and Transcriptomic response.
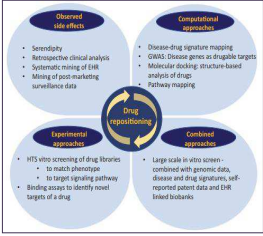
Figure 4: Current and future approaches for drug repositioning.
HTS, high-throughput screening; EHR, electronic health record (Grinnan, Trankle et al. 2019). 1.4 Challenges To Drug Repositioning During drug repositioning, researchers encounter a large number of challenges. Some of these challenges have been described here. 1.4.1 Selection Of An Appropriate Approach Different approaches that may be experimental or computational are used. Selection of an appropriate approach is required (Novac 2013). Many researchers have introduced mixed approaches, in which both computational and experimental approaches are included (Ai et al., 2015; Amelio et al., 2014; Lee et al., 2016; Xue, Li et al., 2018). During mixed approaches, outcomes of computational methods are confirmed by clinical tests and experiments. 1.4.2 Selection Of Target Population Target group selection is an important task in drug repositioning. Incorrect selection of target population results in outcomes other than expected one (Matthews and Mccoy, 2003; Gns, Gr et al. 2019). For example, thalidomide was developed to treat morning sickness in pregnant women but it was found to treat multiple myeloma when this group was removed (Novac, 2013). 1.4.3 Intellectual Property (IP) Protection Intellectual property (IP) issues are issues related to the commercialization. If the drug candidate has never approved for marketing, IP can enhance value of repositioner. However, repositioned candidate already exists in the scientific community, pre-existing patents might block the commercialization of candidate. To defend drugs against competitors, two general cases are considered. First is that composition of matter (COM) IP is held by other party and deals to license. Second case is that compound is off patent. In this case, the repositioner depends on method of use (MOU) patent or new substantial barriers are provided using new drug formulations, dosage forms or drug combinations (Ashburn and Thor 2004). 1.4.4 Overlapping Of Models For drug repositioning, different models must be used. Because overlapping of models results in overlapping of investment issues. 1.4.5 Achievement Within Timeline When an old drug is repositioned for a new indication, appropriate dosage of drug and route of administration must be chosen. Drug formulation is optimized in such a way that it does not destabilize drug (Novac 2013). 1.4.6 Unexpected Adverse Events Drug repositioning involves different groups of patients. Each group has a characteristic set of physiological conditions so each group might 2. Synergistic Drug Repositioning Synergistic drug repositioning refers to identifying new indications for already approved drugs when used in combination. Since the past five years, the number of repurposed drugs has increased. However, many newly developed drug candidates have low potential and many diseases involves various mechanisms. So, combination of two or more drugs is best approach to increase drug potency and to target multiple components of a mechanism. 2.1 Potential For Drug–Drug Interaction During synergistic drug repositioning, the potential of adverse drug-drug interactions (DDI) must be considered. Pharmacodynamics (PD) and pharmacokinetics (PK) are adverse DDIs (Sun, Sanderson et al. 2016). In 1990s, selective serotonin reuptake inhibitor (SSRI) paroxetine was prescribed to treat depression and side effects of tamoxifen in patients of breast cancer. Paroxetine inhibited the metabolism of tamoxifen. A study in 2010 reported that this DDI resulted in increased death rate (Kelly et al., 2010). Therefore, much more care is required during synergistic drug repurposing. 2.2 Approaches Of Synergistic Drug Repositioning Many computational models including bioinformatics and other databases are useful in synergistic drug repositioning. NowakSliwinska et al., (2016) introduced a feedback system control (FSC) to screen various drug combinations. In this method, dose–response curves are plotted after phenotypic cell viability assay for each drug and a differential evolution (DE) algorithm predicts drug combination. Low Daugherty et al., (2016) suggested that to discover drug pairs with synergistic repurposing potential, use of electronic health records (EHRs) is also very effective. 2.3. Success Stories Of Synergistic Drug Repositioning Efficient synergistic drug repositioning resulted in the understanding of complex disease physio-pathology and designing of better treatments. Synergistic drug repositioning has been applied to treat cancer, microbial infections, neurological disorders, diabetes and a large number of rare diseases. Synergistic drug repositioning approach is effective to discover more drug candidates against infectious agents (viruses, bacteria, protozoans and etc.), neurological disorders and cancer.
References
- 1. Al-Mosawi AJ. (2021),. Iraq healthcare system: The first year of covid-19 pandemic. Journal of Clinical and Laboratory Research (ISSN: 2768-0487)17 September; 3(4):1-18. Doi:10.31579/2768-0487/042.
View at Publisher | View at Google Scholar - Alfedi, G., et al. (2019).
View at Publisher | View at Google Scholar - Amelio I, Gostev M, Knight RA, Willis AE, Melino G, Antonov AV. DRUGSURV: a resource for repositioning of approved and experimental drugs in oncology based on patient survival information. Cell Death Dis. 2014;5:e1051
View at Publisher | View at Google Scholar - Ashburn, T. T. and K. B. Thor (2004).
View at Publisher | View at Google Scholar - Brueggeman, L., et al. (2019).
View at Publisher | View at Google Scholar - Deotarse P. P.1, Jain A. S.1, Baile. M. B, et al. Drug repositioning: a review. Int J Pharma Res Rev. 2015; 4:51-58
View at Publisher | View at Google Scholar - de Mello, C. P. P., et al. (2018).
View at Publisher | View at Google Scholar - Fleisher GR, Wilmott CM, Campos JM (1983). Amoxicillin combined with clavulanic acid for the treatment of soft tissue infections in children. Antimicrob Agents Chemother 24: 679–681
View at Publisher | View at Google Scholar - Gerits, E., Defraine, V., Vandamme, K., De Cremer, K., De Brucker, K., Thevissen, K., et al. (2017). Repurposing toremifene for treatment of oral bacterial infections. Antimicrob. Agents Chemother. 61, e1846–e1816. doi: 10. 1128/AAC.01846- 16 Gns, H. S., et al. (2019).
View at Publisher | View at Google Scholar - Graziano TS, Cuzzullin MC, Franco GC, et al. Statins and antimicrobial effects: simvastatin as a potential drug against Staphylococcus aureus biofilm. PLOS ONE. 2015, http://dx.doi.org/10.1371/journal.pone.012 8098.
View at Publisher | View at Google Scholar - Grinnan, D., et al. (2019).
View at Publisher | View at Google Scholar - Han, K., et al. (2017).
View at Publisher | View at Google Scholar - Houang ET, Watson C, Howell R, Chapman M (1984). Ampicillin combined with sulbactam or metronidazole for single-dose chemoprophylaxis in major gynaecological surgery. J Antimicrob Chemother 14: 529– 535.
View at Publisher | View at Google Scholar - KalantarMotamedi, Y., et al. (2018).
View at Publisher | View at Google Scholar - Kelly, C.M. et al. (2010) Selective serotonin reuptake inhibitors and breast cancer mortality in women receiving tamoxifen: a population based cohort study. BMJ 340, c693
View at Publisher | View at Google Scholar - Kola,I. and Landis,J. (2004) Can the pharmaceutical industry reduce attrition rates? Nat. Rev. Drug Discov., 3, 711–715.
View at Publisher | View at Google Scholar - Kondo, T., et al. (2017).
View at Publisher | View at Google Scholar - Kulesa, A., et al. (2018).
View at Publisher | View at Google Scholar

 Clinic
Clinic
