Research Article | DOI: https://doi.org/10.31579/2834-5177/028
Antibiotics Resistance Pattern of Aerobic Bacteria Causing Surgical Site Wound Infection and its Associated Factors in Public Hospital, Dire Dawa-Eastern Ethiopia
1 Department of Medical Laboratory Science, College of Medicine and Health Sciences, Dire Dawa University, Ethiopia
2 School of Medicine, College of Medicine and Health Sciences, Dire Dawa University, Ethiopia
3 Department of Public Health, College of Medicine and Health Sciences, Dire Dawa University, Ethiopia.
*Corresponding Author: Robel Mekonnen Yimer, Department of Medical Laboratory Science, College of Medicine and Health Sciences, Dire Dawa University, Ethiopia.
Citation: Tameru M. Melaku, Robel M. Yimer, Mahad M. Abdinasir, Mesfine K. Alemu, (2023), Antibiotics Resistance Pattern of Aerobic Bacteria Causing Surgical Site Wound Infection and its Associated Factors in Public Hospital, Dire Dawa-Eastern Ethiopia, International Journal of Clinical Infectious Diseases, 2(6); DOI:10.31579/2834-5177/028
Copyright: © 2023, Robel Mekonnen Yimer. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received: 02 May 2023 | Accepted: 14 December 2023 | Published: 29 December 2023
Keywords: carbaryl; protein, albumin; globulin; broiler chicks
Abstract
Purpose: This study aimed to investigate antimicrobial resistance patterns of major bacterial agents isolated from surgical site infections (SSIs) and its associated factors among patients admitted at surgical and gynecology ward of Public Hospital in Dire-Dawa, Eastern Ethiopia.
Methods: A hospital based cross-sectional study was conducted between December 2020 and March, 2021. A total of 188 wound samples along with demographic and clinical data were collected from the study subjects. Isolation and identification of bacteria and antimicrobial susceptibility test (AST) was carried out in microbiology laboratory. The finding from AST was interpreted based on the CLSI guideline. Data were entered and analyzed on SPSS V.24. Descriptive statistics and Binary logistic regression analysis were computed to present the findings with a p-value < 0.05 set as level of significance.
Results: The prevalence of culture positive SSIs found to be 59.1% (95% CI: 52.1-66.1). A total of 120 bacteria were identified; S. aureus 43(35.8%), GB Streptococci spp 20(16.7%), and E. coli 16(13.3%) were frequently isolated bacteria. Gram positive bacteria showed maximum resistance to Co-trimoxazole, Penicillin, Clindamycin, and Chloramphenicol. By the same token, Gram negative bacteria showed maximum resistance to Ampicillin, Cloxacilin, and Cefepime; while lower resistance to Vancomycin, Ceftriaxone and Gentamicin. Multi-drug resistance (MDR) levels were determined to be 85% overall. Alcohol consumption, female gender, the type of wound, and the type of wards were all strongly linked to bacterial SSIs. (p-value < 0.05).
Conclusions: The studied area has an extremely high level of MDR as shown by the SSI bacterial isolate. The fact that clinicians have few drug options to treat patients with SSIs raises serious medical concerns. Therefore, it is strongly advised to use antimicrobial agents wisely and practice aseptic procedures to reduce the establishment of antibiotic resistance.
Introduction
A wound is a tear in the skin and the exposing of subcutaneous tissue when the skin's integrity has been lost.1 A variety of bacteria can colonize and contaminate the exposed subcutaneous tissues, and the conditions for microbial development become ideal if the affected tissue becomes devitalized and the host immune system is weakened. This is due to the fact that the host immune response is crucial in determining whether or not wound infections will develop.2 Various types of microbes, including bacteria, fungi, and parasites, can infect a wound.
Surgical site wound infection (SSI) is the most costly post-operative complication and also the second-most prevalent nosocomial infections (NIs).3 In most case, SSI occurs during one month after surgical operation or one year after implant surgery.4 Sixty to eighty percent of SSIs arise in the incision; a minority involve deep soft tissue or surrounding areas.5 Around 25% of all NIs and over one-third of postoperative deaths have been documented to be related to SSIs.6 According to several studies, people with SSI experience longer hospital stays and more financial burdens than those without the infection who underwent comparable surgical procedures.6-8
The occurrence of SSI is determined by the site of wound contamination and the virulence/ pathogenicity of the microorganisms, in balance with the host's immune response.9 Additionally, incidence of SSIs varies greatly between surgical procedures, hospitals, patients, among surgeons, the surveillance criteria used, and the quality of data collection.10
Staphylococcus aureus is regarded as a significant human pathogen and the leading cause of SSIs globally, with a prevalence rate ranging from 4.6 to 54.4 percent.10 In Ethiopia, different studies have indicated that the prevalence of SSI ranges from 11.8%-84.1%.11-13 S. aureus, Kelbsiella species, E. coli, Proteus species, Streptococcus species, Enterobacter species, Pseudomonas species and Coagulase negative Staphylococci (GoNS) reported as the most common pathogens.14-15 However, the location of the wound, the patients' exposure, and the hygienic conditions of the hospital will determine which organism(s) are at play.16
The problem of SSI has, more than ever, taken a new dimension as a result of multi-drug resistant organisms (MDROs): SSIs brought on by MDROs significantly increases patient morbidity, mortality, and incur considerable healthcare costs.17 Antibiotics are frequently bought without a prescription in Ethiopia and the majority of developing nations. This results in widespread public antibiotic abuse, which fuels the emergence and spread of antimicrobial resistance.18-19 Due to extensive drug resistant bacterial strain, a rise in infections brought on by methicillin-resistant S. aureus (MRSA), and polymicrobic flora, controlling wound infections has become more difficult.20 Therefore, if affected patients are to receive high-quality medical care, routine bacteriological evaluation of infected wounds is essential, especially when blind treatment is required, as it often is in developing and/or underdeveloped countries.
Little is currently known about the frequency of SSIs caused by MDROs in Ethiopia. There have been no prior studies on the predictors of SSIs, particularly in the Eastern region of Ethiopia, which includes Dire Dawa, Harari and Somali. There is also no well-established epidemiological data on AST. Since the magnitude of drug resistant bacteria significantly differs by geography and hospital settings, and microorganisms may acquire resistance to antibiotics over time, current knowledge on the magnitude of SSIs due to MDROs and up-to-date data regarding antibiotic resistance profile is of the highest importance. This current study investigated bacterial agents of SSIs, their antibiotic resistance profile, and associated factors in Dil-chora referral hospital (DCRH), Dire Dawa, Ethiopia. The information obtained from AST can be used 1) to guide clinicians with the best drug option for patient treatment 2) to keep track on changing drug resistant bacteria that may indicate emergence or transmission of MDROs 3) as baseline data for implementing targeted infection prevention measures as well as to formulate drug prescription policy at national level.
Materials and methods
Study area
Dil-chora Referral Hospital served as the site of the current study. The hospital is located in Dire Dawa administration, eastern region of Ethiopia. According to the Central Statistics Agency of Ethiopia's population projection for the year 2019/2020, it comprises a total population of 550,000.21
The current study was conducted during the first year of COVID-19 pandemic, a time when the majority of public hospitals served as COVID-19 treatment centers and only Dil-chora Referral Hospital offered routine medical and surgical care to patients from the three regions in the eastern part of Ethiopia.
Study design and period
Hospital based cross sectional study was conducted between December, 2020 and March, 2021 at Public Hospital in Dire Dawa.
Source and Study population
All patients who underwent surgical procedures at Dil-chora Referral Hospital during the study period comprised the source population. Whereas, patients who had developed SSIs during their hospital stay and/or after they are discharged were the study populations.
Inclusion and exclusion criteria:
During the study period, patients who developed sepsis-related clinical symptoms were included. Patients who had a history of antibiotic treatment one week prior at the time of data collection and those who did not develop wound infection based on clinical examination were excluded.
Sample size determination and sampling technique
The sample size was calculated using single population proportion formula by considering 95% confidence interval, 5% margin of error, and 12.4% prevalence of SSIs taken from previous study in Debre Markos.22
n = z (α/2)2 p (1-p)/d2
Where
n = Sample size
α = level of significance
z = at 95% confidence interval Z value (α = 0.05) = Z α/2 = 1.96
p = Proportion of wound infection to be studied 12.7% (0.127)
d = Margin of error at (5%) (0.05)
Thus n= (1.96)2x 0.124(1-0.124)/ (0.05)2
n= (3.8416) x (0.108624)/ (0.0025)
n= 170.36≈ 171
Adding 10% contingency for non-response rate makes the total sample size n = 171 + 17 = 188
Accordingly, patients with SSIs were selected by systematic sampling and included in the study.
Data collection tool and procedure
A face to face interview and checklist were used to collect socio-demographic variables, clinical history, and possible factors related to surgical wound infection using pre-design questionnaire. The questionnaire was adapted from previous studies and by reviewing different literatures. A physician conducted a clinical examination on patients who were admitted to the surgical and gynecology wards. Wound examination as well as review of procedures that can cause infection, was done so as to identify any underlying risk factors. Attendant Nurses in surgical ward (SW), orthopedic ward, and gynecology ward had collected the primary data after securing written informed consent from all study participants.
Sample Collection and Bacteria Identification
Wound Sample Collection and processing
Laboratory professionals who have undergone training in collecting wound swabs did so while according to SOPs. Swab samples were taken during the time of wound dressing and before the surgical site was treated with an antiseptic solution Levine's approach was used to prepare the wound beds, which involved cleaning the wound surface and contaminants using sterile gauze moistened with sterile normal saline solution. Two wound samples were then taken from the infected area, without contaminating them with skin commensals, immediately placed into Brain Heart Infusion (BHI) transport medium, properly labeled and taken to the microbiological laboratory unit at Ethiopia Public Health Institute (EPHI) for examination.
Isolation and identification of bacterial species
The first wound swab was used to make Gram stain smears. The second wound swab, however, was inoculated into blood agar (Oxoid, Ltd., UK), MacConkey agar (Oxoid, Ltd., UK), and mannitol-salt agar (Oxoid, Ltd., UK). All plates were incubated at 37°C for 24-48 hours. Gram-staining reaction, colony features, and biochemical assays were used to identify positive cultures in accordance with standard bacteriological procedures.23-24 Gram negative bacterial isolates were identified using the triple sugar iron (TSI), sulphur indole motility (SIM), citrate, urease, and oxidase tests. The identification of isolates of Gram-positive bacteria also included testing for the enzymes coagulase and catalase, as well as bacitracin and optochin sensitivity assays.
Antimicrobial susceptibility testing
Antibiotic susceptibility testing (AST) was done by the Kirby-Bauer disc diffusion method on Mueller Hinton (MH) agar (Oxoid Ltd., UK). Three to five colonies from a pure culture were taken, added to a tube containing 5 ml of nutrient broth, and then gently mixed to create a homogeneous suspension. The suspension was incubated at 37°C until the turbidity was brought up to the McFarland standard of 0.5. Using a sterile cotton swab, the colonies were evenly spaced on the surface of MH agar and allowed to dry for 3-5 minutes at room temperature. To ensure a secure adherence, the antibiotic discs were very gently pressed into the surface of the plates using sterile forceps. The plates were then incubated for 24 hours at 37°C. The diameters of the zone of inhibition around the discs were measured to the nearest millimeter using a ruler held on the back of the inverted petriplate. Finally, the tested bacteria were categorized as sensitive, intermediate, or resistant using the standardized table provided by the clinical laboratory standards institute (CLSI).24 Selection of antibiotics was in compliance with the local prescription policy and CLSI guideline.
Quality assurance of data collection and laboratory procedures
The principal investigator gave two days training to data collectors about the questionnaire and data collection techniques. The questionnaire was pre-tested on sample populations not included in the study. Then the collected data was checked for completeness at the end of data collection.
During laboratory analysis, standard operating procedures were strictly followed, calibrated equipment was used for measuring reagents, and all materials were checked for proper functionality. All culture media were prepared following manufacturer's instruction. Then sterility of prepared culture media was checked by incubating 5% of the batch at 35 – 37°C overnight and observed for any bacterial growth. Moreover, Staphylococcus aureus (ATCC-25923), Escherichia coli (ATCC-25922) and Pseudomonas aeruginosa (ATCC-27853) were used as a quality control strains for culture and antimicrobial susceptibility testing.
Statistical analysis
EpiData v.3 used to enter pre-coded data, checked for completeness, and then transported to SPSS V.24 for analysis. Mean, standard deviation, frequency, and percentage were analyzed and summarized in tables. Bivariate and multivariate logistic regression analysis was computed to identify factors associated with occurrence of SSIs. Adjusted odds ratio with 95% confidence interval were used to estimate the strength of the association and p-value <0 xss=removed>
Ethics approval and consent to participate
This study was carried out after receiving institutional review board approval from Dire Dawa University (approval number: SPD 035/0068). Study participants have given an informed written consent after being told of its goal and purpose.
Results
Socio-Demographic characteristics of the study participants
A total of 188 patients clinically diagnosed with SSWI at public hospital were included. The median age of the participants was 26 years. Higher rate of bacterial SSIs observed among patients with age group of 35-49 years and above 50 years at 50(45%) and 37(33.3%), respectively. Similarly, rate of SSIs were higher among females 82(73.9%), urban residents 80(72.1%), those attended primary school 39(35.1%), and private employees 26(23.4%) (Table 1).
Variables | Category | Bacterial SSIs | Total | |
Yes | No | |||
Age group | < 20> | 4(3.6%) | 4(5.2%) | 8(4.3%) |
20-34 | 20(18.0%) | 29(37.7%) | 49(26.1%) | |
35-49 | 50(45.0%) | 27(35.1%) | 77(41.0%) | |
>50 | 37(33.3%) | 17(22.1%) | 54(28.7%) | |
Sex | Male | 29(26.1%) | 43(55.8%) | 72(38.3%) |
Female | 82(73.9%) | 34(44.2%) | 116(61.7%) | |
Residence | Urban | 80(72.1%) | 62(80.5%) | 142(75.5%) |
Rural | 31(27.9%) | 15(19.5%) | 46(24.5%) | |
Educational status | Illiterate | 26(23.4%) | 18(23.4%) | 44(23.4%) |
Primary education | 39(35.1%) | 19(24.7%) | 58(30.9%) | |
Secondary | 27(24.3%) | 16(20.8%) | 43(22.9%) | |
Diploma & above | 19(17.1%) | 24(31.2%) | 43(22.9%) | |
Occupation | Student | 10(9.0%) | 15(19.5%) | 25(13.3%) |
Housewife | 16(14.4%) | 9(11.7%) | 25(13.3%) | |
Farmer | 11(9.9%) | 6(7.8%) | 17(9.0%) | |
Private employee | 26(23.4%) | 16(20.8%) | 42(22.3%) | |
Governmental | 12(10.8%) | 16(20.8%) | 28(14.9%) | |
Others | 36(32.3%) | 15(19.5%) | 51(27.1%) | |
Table 1: Socio-demographic characteristics of patients with surgical site infection at public Hospital in Dire Dawa, Eastern Ethiopia (n=188)
Clinical and Health Related Characteristics
As displayed on Table 2 below, 58(52.4%), 25(22.5%), and 50(45.0%) bacterial SSIs cases were among patients who chew Khat, drink alcohol and smoke cigarette, respectively. More SSIs observed among patients admitted in obstetrics/gynecology ward 55(49.5%), those having had CS surgeries 71(64.0%); whereas 44(39.6%) SSIs cases detected among patients with > 7 days LOS. Almost equal proportion of cases were observed for elective 55(49.5%) and emergency surgery 56(50.5%). Patients with DM comorbidity showed higher SSIs cases at 95(85.6%). On the contrary, 76(68.5%) of cases were detected from clean wounds and 59(53.2%) from those with post-operative prophylaxis (Table 2).
Variables | Category | Bacterial SSIs | Total | |
Yes | No | |||
Khat chewing | Yes | 58(52.3%) | 20(26.0%) | 78(41.5%) |
No | 53(47.7%) | 57(74.0%) | 110(58.5%) | |
Drink alcohol | Yes | 25(22.5%) | 41(53.2%) | 66(35.1%) |
No | 86(77.5%) | 36(46.8%) | 122(64.9%) | |
Smoke cigarette | Yes | 50(45.0%) | 35(45.5%) | 85(45.2%) |
No | 61(55.0%) | 42(54.5%) | 103(54.8%) | |
Type of ward | Gyny | 55(49.5%) | 21(27.3%) | 76(40.4%) |
Surgical | 11(9.9%) | 36(46.8%) | 47(25.0%) | |
Orthopedics | 45(40.5%) | 20(26.0%) | 65(34.6%) | |
Type of surgery | Appendectomy | 7(6.3%) | 17(22.1%) | 24(12.8%) |
Mastectomy | 16(14.4%) | 19(24.7%) | 35(18.6%) | |
CS | 71(64.0%) | 22(28.6%) | 93(49.5%) | |
Debridement | 17(15.3%) | 19(24.7%) | 36(19.1%) | |
Duration of stay | > 7 days | 44(39.6%) | 51(66.2%) | 95(50.5%) |
< 7> | 67(60.4%) | 26(33.8%) | 93(49.5%) | |
Schedule of surgery | Elective | 55(49.5%) | 66(85.7%) | 121(64.4%) |
Emergency | 56(50.5%) | 11(14.3%) | 67(35.6%) | |
Comorbidity | DM | 95(85.6%) | 19(24.7%) | 114(60.6%) |
Anemia | 4(3.6%) | 43(55.8%) | 47(25.0%) | |
bleeding disorder | 1(.9%) | 3(3.9%) | 4(2.1%) | |
Others | 11(9.9%) | 12(15.6%) | 23(12.2%) | |
Type of wound/operation | Clean | 76(68.5%) | 39(50.6%) | 115(61.2%) |
Contaminated | 25(22.5%) | 17(22.1%) | 42(22.3%) | |
Dirty | 10(9.0%) | 21(27.3%) | 31(16.5%) | |
SAP | Preoperative | 52(46.8%) | 44(57.1%) | 96(51.1%) |
Post-operative | 59(53.2%) | 22(28.6%) | 81(43.1%) | |
Neither | 0(.0%) | 11(14.3%) | 11(5.9%) | |
Abbreviation: CS, Cesarean Section; DM, Diabetes Mellitus; SAP, Surgical Antibiotic Prophylaxis;
Table 2: Clinical and Health Related Characteristic of patients with surgical site infection at public Hospital in Dire Dawa, Eastern Ethiopia (n=188)
Proportion of Bacteria Isolated from patients with SSIs
Out of the total 188 wound swabs/pus aspirates collected from patients clinically diagnosed with SSIs at Dil-chora Referral Hospital, 111 samples showed bacterial growth with overall prevalence of culture confirmed SSIs 59.1% (95% CI: 52.1 – 66.1), yielding 120 bacterial species. Out of these, 102(91.9%) infections were due to single bacterial agents while it was 9(8.1%) for double infections. Gram-positive bacteria were the major isolate at 68(56.7%), the predominant being S. aureus 43(35.8%), followed by GB Streptococci spp 20(16.7%), and the least isolate was Enterococcus spp at 5(4.2%). On the other hand, from a total of 52(43.3%) Gram negative bacterial isolates, both E. coli and K. pneumoniae predominate at 16(13.3%), followed by P. aeruginosa 9(7.5%), P. mirabilis 7(5.8%), and Citrobacter was the least isolated at 4(3.3%) (Figure -1).
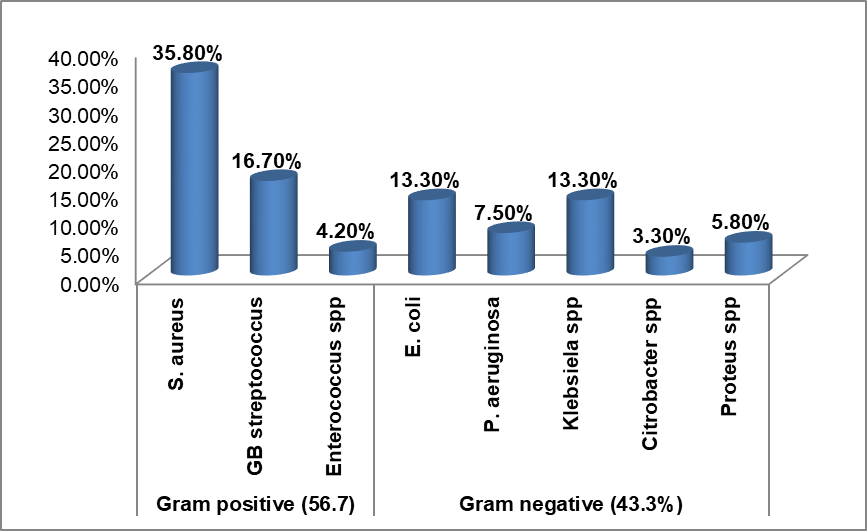
Figure 1: Proportion of bacterial species isolated from surgical site wound infection at public hospitals in Dire Dawa, Eastern Ethiopia.
Antimicrobial resistance pattern of bacteria to individual drug
Generally, Gram positive bacteria showed maximum resistance of above 75% for Tetracycline, Cotrimoxazole, Penicillin, Clindamicin, and Chloramphenicol but lower 25.0% and 29.4% for Gentamicin and Vancomycin. Similarly Gram negative showed maximum resistance of above 70% for Ampicillin, Cloxacilin, Cotrimoxazole, Oxacillin, and Cefepime; while only 46.2% and 30.8% for Ceftriaxone and Gentamicin, respectively. More specifically, S. aureus showed 100% resistance against Clindamycin; 90.7% resistance to Penicillin, Co-trimoxazole, and Chloramphenicol; 65.1% against Azithromycin and 51.2% for Oxacilin. Low level of resistance was conferred to Gentamicin and Vancomycin at 34.9% and 20.9%, respectively. Similarly, Enterococcus spp showed 100% resistance against Penicillin and Cotrimoxazole, 80% to Tetracycline and Vancomycin, while 60
Discussion
Surgical site wound infection is the major cause of morbidity and mortality following surgery.11 The emergence of antibiotic-resistant bacteria is known to cause major consequences for public health.2 Patients with SWI brought on by microorganisms that are resistant to antibiotics run the risk of having worse clinical outcomes and using up more resources in the healthcare system than their counterparts/peers.
In this study the prevalence of culture confirmed SSIs found to be 59.1% which is in line with earlier research report from Nepal (64.5%), Uganda (58.5%), and Nigeria (64.8%).20,25-26 On the other hand, our finding was higher than a report from Harar town (11.8%) and Hawassa (24.6%)12-13, other country in Rwanda (10.2%), Uganda (15.5%), and Peru (2.5%).6,27-28 However, higher prevalence of SSIs was reported by others.14,29-31 Differences in the distribution of nosocomial pathogens, surgical methods, and operating rooms, shortage of trained personnel, failure to maintain sterility during surgical procedures, and insufficient infection prevention & control practices among countries and health facilities, could be the main reasons for observed discrepancies in the proportion of SSI by these studies.
In the current study, 91.9% SSIs were due to single bacterial agents while it was 8.1% for double infections. This finding is consistent with similar studies done in Ethiopia and abroad.29,32-33 Other studies, however, reported higher rate of mixed infection in Jimma and Nigeria.29,34
According to our study, S. aureus was the most frequently isolated bacteria which disagree with the findings of several previous studies where E. coli was the dominant bacteria.29,35-36 Whereas K. pneumonia was the commonest isolates (50%) according to the study in Uganda.27 The isolation rate of S. aureus (35.8%) in our study was comparable with the finding of the study carried out in Harar (30%), Addis Ababa (33.3%), and Bahir Dar (26.2%).12,15,36 The observed discrepancy could be explained by geographical difference and/or difference in prevalent bacterial flora by hospital settings.
The current study also identified diverse antimicrobial susceptibility patterns of the bacterial isolate with the various tested drugs. S. aureus demonstrated higher level of resistance (100%) against Clindamycin; 90.7% resistance to Penicillin, Cotrimoxazole, and Chloramphenicol; and 65.1% against Azithromicin. This finding was comparable with several studies done at different times, such as in Gondor, Addis Abeba Black lion Hospital, and Uganda.13,15 Lower level resistance conferred by S. aureus to Gentamicin (34.9%) and Vancomycin (20.9%) in our study contradicts with other study. For instance, high sensitive (100%) for Vancomicin reported by the studies in India and Nepal,37-38 in addition to 89.7% and 80% sensitive reported for Gentamicin in India and Jimma.37,39
Among all isolates of S. aureus, 51.2% of them found to be MRSA based on disc duffusion test for Oxacilin. This finding was higher compared to reported by Suchitra (42%), the study in India (20%) and in Nepal 2013 (41.7%), and Adwan et al (33.3%).24,37-38,40 On the contrary, our finding is lower compared to reported in Western Nepal (60%) and Jimma (76.7%).20,39 Enterococcus spp showed 100% resistance to Pencillin and Co-trimoxazole, 80% for Vancomycin which is higher compared to other studies.14,37,40
Among Gram negative bacteria, E. coli demonstrated 93.7% resistance to Ampicillin which is higher than reported by previous studies.14,29,37 Similarly, 81.2% of E. coli resisted Cefepime and it was 66.7% in Jimma.29 Higher resistance of E. coli against Gentamicin, Ceftriaxone, and Cefotaxim was reported in Jimma and Gondar,14,29 in contrary to the current study where lower resistance rate of 12.5%, 18.8%, and 37.5% observed for Gentamicin, Ceftriaxone, and Cefotaxim, respectively.
P. aeruginosa showed 100% resistance against Ampicillin and Cefotaxime, and 88.9% to Cefepime which is higher compared to reported resistance of 98.2% to Ampicillin in Western Nepal,20 54% to cefotaxime by Suchitra,24 and 62.5% to Cefepime in India.37 Although 77.8% and 44.4% of P. aeruginosa in our study resisted Ceftriaxone and Ciprofloxacin, relatively higher resistance rate for both drugs were reported in Western Nepal and in India.20,37
In the current study, all Klebsiella spp conferred 100% resistance against Cotrimoxazole and Cefepime which is higher compared to study in India (54.4%).37 On the other hand, 62.5% and 31.2% resistance rate of Klebsiella spp to Ampicillin and Gentamycin was lower compared to reported 100% and 79.0% resistance to Ampicillin in Gondar and India.14,37
According to previous study, Citrobacter and Proteus spp showed 100% resistance to Ampicillin and Cefepime, respectively 14,29 which is in agreement with the present study. However, the study in Gondar indicated that all Proteus spp were 100% resistant against Ampicillin, Gentamycin, and Ceftazidime,14 which is higher compared to the current study finding. This could be due to difference in the number of total isolates where only one Proteus spp was isolated and tested in the other study. Likewise, resistance of Citrobacter to Ceftriaxone and Ceftazidime (75%) reported in Jimma Medical Center is also higher compared to the present study (25%).29
Our study revealed that 85.0% of the bacteria isolated from SSIs found to be multi-drug resistant (MDR), which is consistent with study findings in Ethiopia and Nepal.20,30 On the other hand, although lower level of MDR (39.2%) was reported by Chaudhary et al 41 other studies, however, reported greater MDR percentage.20,6
More specifically, Gram positive bacteria showed higher MDR level at 86.7% relative to Gram negative (82.7%). This finding is comparably higher than reported in Gondar and Jimma.2,39 Moreover, the current study revealed that all Enterococcus Spp., Citrobacter species, P. aeruginosa, and Proteus spp exhibited 100% level of MDR. Our finding is higher compared to reported by Godebo et al.39 The possible reasons for the observed higher resistance to these drugs in the current study is that majority of the listed antimicrobials are widely accessible at neighborhood pharmacies and are usable by anyone without a prescription. This would contribute to the emergence and spread of antibiotic resistance in this study area.
The current study provides ample evidence to suggest that bacteria flora within hospital environments are becoming alarmingly drug- resistant and the problem gets even worse when they cause HAIs, such as SSIs. In the study area, where there are lax regulatory practices and insufficient bacteriological surveillance due to a lack of facilities for routine AST, empiric treatment, overuse, and misuse of antibiotics may all be contributing factors to the reported increase in MDR. Previous study conducted on the same hospital setting suggested the importance of monitoring microbial air contamination by providing ample evidence that potential pathogenic bacteria carried on indoor air as well as surfaces of patient caring equipment are the major source of HAIs and spread of MDROs.42
On binary logistic regression analysis, the present study revealed that female sex, drinking alcohol, contaminated and dirty wound, and type of wards (SW), all have identified as factors associated with incidence of SSI. A similar study in Rwanda supported our finding where SSI incidence was higher in contaminated and dirty wounds, and long hospital stay of more than 14 days.6 Similarly, Fisha et al reported that clean-contaminated and dirty wounds and length of hospital stay≥6 days increased SSIs.8 Moreover, according to Adwan et al, female (p=0.070) and type of surgeries were the risk factors associated with SSIs.40
Limitation
This current study did not isolate strict anaerobic bacteria and fungal agents of SSI. We suggest other researchers to further investigate.
Conclusion
The studied area has an extremely high level of MDR as shown by the SSI bacterial isolate. The three dominant bacteria were S. aureus, GB Streptococci, and E. coli. The fact that clinicians have few drug options to treat patients with SSIs raises serious medical concerns. Gentamycin, vancomycin, and ciprofloxacin, on the other hand, can be recommended as efficient medications against Gram positive and Gram negative bacteria and should be used as drug of preference in the management of SSIs at the hospital.
Generally, female gender, alcohol consumption, contaminated and dirty wound, and type of wards (SW), are all identified as factors associated with incidence of SSI. Community level health education campaign targeted at behavioral modification is essential.
Recommendation
- Practicing aseptic procedures and rational use of antimicrobial agents leading to minimize infection rate and emergence of drug resistance is mandatory.
- Following recommended cleaning, disinfection and sterilization procedures for maintaining patient care areas and equipment is essential to avert patient morbidity and mortality by SSI due to resistant bacteria.
- Conduct periodic surveillance of antimicrobial resistance pattern of bacterial agents of SSIs to limit further emergence and spread among bacterial pathogens.
- We recommend other researchers to further investigate molecular typing of the bacterial isolates to confirm transmission or delineate epidemiology of MDRO in facility.
- Acknowledgements
We would like to acknowledge Dire Dawa University, Dil-chora referral hospital directors, data collectors, and most importantly, the study participants.
Disclosure of conflict of interest
The author(s) declared no potential conflict of interest with respect to the research, authorship, and/or publication of this paper.
Funding
This research has never received financial supported from any funders.
References
- Nitin Goel Insan, Nikhil Payal, Mahesh Singh, Amod Yadav, B.L Chaudhary. Post operative wound infection: Bacteriology and antibiotic sensitivity pattern. Int J Cur Res Rev. 2013; 05(13):74-79.
View at Publisher | View at Google Scholar - D. Amenu, T. Belachew, and F. Araya, “Surgical site infection rate and risk factors among obstetric cases of Jimma University Specialized Hospital, Southwest Ethiopia,” Ethiopian Journal of Health Sciences. 2011; 21(2): 91–100.
View at Publisher | View at Google Scholar - Dessie w, Mulugeta G, Fentaw S, Mihret A, Hassen M, Abebe E. Pattern of Bacterial Pathogens and Their Susceptibility Isolated from Surgical Site Infections at Selected Referral Hospitals, Addis Ababa, Ethiopia. International Journal of Microbiology. 2016; Article ID 2418902.
View at Publisher | View at Google Scholar - Horan TC, Gaynes RP, Martone WJ, Jarvis WR, Emori TG. CDC definitions of noscomial surgical site infections, 1992: a modification of CDC definitions of surgical wound infections. Infect Control Hosp Epidemiol. 1992;13:606-8.
View at Publisher | View at Google Scholar - Wong E S. Surgical site infections. In: Mayhall CG, editor. Hospital epidemiology and infection control. 1st ed.U.S.A: Williams and Wilkins; 1996.p.154-74.
View at Publisher | View at Google Scholar - Mukagendaneza, M.J., Munyaneza, E., Muhawenayo, E., Nyirasebura, D., Abahuje, E., Nyirigira,J. et al. Incidence, root causes, and outcomes of surgical site infections in a tertiary care hospital in Rwanda: a prospective observational cohort study. Patient Saf Surg. 2019; 13(1):1-8 https://doi.org/10.1186/s13037-019-0190-8
View at Publisher | View at Google Scholar - Reichman D E, Greenberg J A. Reducing Surgical Site Infections: A Review. Rev Obstet Gynecol. 2009; 2:212-21.
View at Publisher | View at Google Scholar - Fisha K, Agage M, Mulat G and Tamrat KS. The prevalence and root causes of surgical site infections in public versus private hospitals in Ethiopia: a retrospective observational cohort study. Patient Saf Surg 2019;13: 26. https://doi.org/10.1186/s13037- 019-0206-4
View at Publisher | View at Google Scholar - Barie PS. Surgical site infections: epidemiology and prevention. Surg Infect. 2002;3(S1):s9–21.
View at Publisher | View at Google Scholar - Owens CD, Stoessel K. Surgical site infections: epidemiology, microbiology and prevention. J Hosp Infect. 2008; 70 Suppl 2:3-10.
View at Publisher | View at Google Scholar - Dagnachew Muluye, Yitayih Wondimeneh, et.al Bacterial isolates and their antibiotic susceptibility patterns among patients with pus and/or wound discharge at Gondar university hospital. BMC Research Notes. 2014; 7:619
View at Publisher | View at Google Scholar - Anwar Shakir, Degu Abate,Fikru Tebeje et al, Magnitude of Surgical Site Infections, Bacterial Etiologies, Associated Factors and Antimicrobial Susceptibility Patterns of Isolates Among Post-Operative Patients in Harari Region Public Hospitals, Harar, Eastern Ethiopia. Dovepress Infection and Drug Resistance 2021:14 4629–4639
View at Publisher | View at Google Scholar - Deribe B, Jemebere W, Bekele G Surgical site infection prevalence and associated factors in Hawassa University comprehensive specialized hospital, southern Ethiopia; 2019. doi:10.21203/rs.2.15549
View at Publisher | View at Google Scholar - Kifilie AB, Dagnew M, Tegenie B, Yeshitela B, Howe R, Abate E. Bacterial Profile, Antibacterial Resistance Pattern, and Associated Factors from Women Attending Postnatal Health Service at University of Gondar Teaching Hospital, Northwest Ethiopia.International of Microbiology. 2018; Article ID 3165391, https://doi.org/10.1155/2018/316539
View at Publisher | View at Google Scholar - Asres GS, Legese MH, Woldearegay GM. Prevalence of multidrug resistant bacteria in postoperative wound infections at Tikur Anbessa Specialized Hospital, Addis Ababa, Ethiopia. Arch Med. 2017;9(4):12.(6studies)
View at Publisher | View at Google Scholar - Adegoke, Anthony A, Tom Mvuyo, Okoh Anthony I. and Jacob Steve. Studies on multiple antibiotic resistant bacteria isolated from surgical site infection. Scientific Research and Essays.2010;5(24):3876-3881.
View at Publisher | View at Google Scholar - Harding K, Renyi R. The International Wound Infection Institute – a new global platform for the clinical management of infected wounds. Int Wound J. 2009; 6(3): 175 78.
View at Publisher | View at Google Scholar - Amare B, Abdurrahman Z, Moges B, Ali J, Muluken L, Alemayehu M, Yifru S et al. Postoperative surgical site bacterial infections and drug susceptibility patterns at Gondar University Teaching Hospital, Northwest Ethiopia. J Bacteriol Parasitol.2011; 2(8): 126.
View at Publisher | View at Google Scholar - Khatiwada S, Acharya S, Poudel R, Raut S, Khanal R, Karna SL, Poudel A. (2020),.Antibiotics Sensitivity Pattern of Post-Operative Wound Infections in a Tertiary Care Hospital, Western Nepal. Preprint from Research Square, Central Statistics Agency, Ethiopia 2007.
View at Publisher | View at Google Scholar - Hulubante BizuayewI, Haimanot Abebe et.al (2020),.Post-cesarean section surgical site infection and associated factors in East Gojjam zone primary hospitals, Amhara region, North West Ethiopia,
View at Publisher | View at Google Scholar - Cheesbrough M. District laboratory practice in tropical countries volume II: microbiology. Cambridge (UK). Page 26-196
View at Publisher | View at Google Scholar - Suchitra Joyce B. and Lakshmidevi N. (2009),.Surgical site infections: Assessing risk factors, outcomes and antimicrobial sensitivity patterns . African Journal of Microbiology Research. ; Vol. 3 (4): 175-179
View at Publisher | View at Google Scholar - JR Anguzu and D Olila . (2007),.Drug sensitivity patterns of bacterial isolates from septic post-operative wounds in a regional referral hospital in Uganda. Afr Health Sci. ; 7(3): 148–154.
View at Publisher | View at Google Scholar - Egbe CA, Omoregie R, Igbarumah IO, Onemu S. (2011),.Microbiology of wound infections and its associated risk factors among patients of a Tertiary hospital in Benin City, Nigeria. JRHS. ;11(2):109-113
View at Publisher | View at Google Scholar - Lubega A., Joel B., and Justina Lucy N., (2017),.Incidence and etiology of surgical site infections among emer gency postoperative patients in mbarara regional referral hospital, South Western Uganda. Surgery research and practice, 2017:1–6.
View at Publisher | View at Google Scholar - Ramirez-Wong FM, Atencio-Espinoza T, Rosenthal VD, et al. (2015),.Surgical site infections rates in more than 13,000 surgical procedures in three cities in Peru: findings of the international nosocomial infection control consortium. Surg Infect. ;16(5):572–576.
View at Publisher | View at Google Scholar - Gemedo Misha, Legese Chelkeba and Tsegaye Melaku. (2021),.Bacterial profile and antimicrobial susceptibility patterns of isolates among patients diagnosed with surgical site infection at a tertiary teaching hospital in Ethiopia: a prospective cohort study Ann Clin Microbiol Antimicrob. ; 20(33): 1-10.
View at Publisher | View at Google Scholar - Mengesha RE, et al. (2014),.Aerobic bacteria in post surgical wound infections and pattern of their antimicrobial susceptibility in Ayder Teaching and Referral Hospital, Mekelle, Ethiopia. BMC Res Notes. 2014;7(1):1–6.
View at Publisher | View at Google Scholar - Zahran WA, Zein-Eldeen AA, Hamam SS, Sabal MSE. (2017),.Surgical site infections: problem of multidrug-resistant bacteria. Menoufia Med J. 2017;30(4):1005–1013.
View at Publisher | View at Google Scholar - Gelaw, S. Gebre Selassie, M. Tiruneh, and M. Fentie, (2013 ),.“Antimicrobial susceptibility patterns of bacterial isolates from patients with postoperative surgical site infection, health professionals and environmental samples at a tertiary level hospital, North West Ethiopia,” International Journal of Pharmaceutical Sciences Review and Research, vol. 3, no. 1, pp. 1–9,.
View at Publisher | View at Google Scholar - Amare, Z. Abdurrahman, B. Moges, and J. Ali, (2011 ),.“Postoperative surgical site bacterial infections and drug susceptibility patterns at Gondar University Teaching Hospital, Northwest Ethiopia,” Journal of Bacteriology & Parasitology, vol. 2, no. 8, 2011.
View at Publisher | View at Google Scholar - Kumalo A, Kassa T, S Mariam Z, Deresse Daka D, Tadesse AH. (2016),Bacterial profile of adult sepsis and their antimicrobial susceptibility pattern at Jimma University Specialized Hospital, South West Ethiopia. Health Sci J. 2016;10(2):3
View at Publisher | View at Google Scholar - Gupta S, Kashyap B. Bacteriological profile and antibiogram of blood culture isolates from a tertiary care hospital of North India. Trop J Med Res. 2016;19:94–99.
View at Publisher | View at Google Scholar - Mulu W, Kibru G, Beyene G, Damtie H. (2013 ),Associated risk factors for postoperative nosocomial infections among patients admitted at Felege Hiwot Referral Hospital, Bahir Dar,Northwest Ethiopia. Clin Med Res. ;2(6):140–147. doi:10.11648/j.cmr.20130206.15
View at Publisher | View at Google Scholar - Anusuya Devi Devaraju and S. Latha Roy. (2021),.Antimicrobial Susceptibility Pattern of Bacterial Isolates from Surgical Site Infections in a Tertiary Care Hospital. Int. J. Curr. Microbiol. App. Sci. ; 10(11): 369-375. https://doi.org/10.20546/ijcmas.2021.1011.042
View at Publisher | View at Google Scholar - M. Raza, A. Chander and A. Ranabhat, (2003 ),.
View at Publisher | View at Google Scholar - Girma Godebo, Gebre Kibru and Himanot Tassew. (2013 ),.Multidrug-resistant bacteria isolates in infected wounds at Jimma University Specialized Hospital, Ethiopia. Annals of Clinical Microbiology and Antimicrobials.;12(17):1-7
View at Publisher | View at Google Scholar - Ghaleb Adwan, Nael Abu Hasan, Ibrahim Sabra, Dalia Sabra, Shorouq Al-butmah, Shorouq Odeh et al. (2016),.Detection of bacterial pathogens in surgical site infections and their antibiotic sensitivity profile. International Journal of Medical Research & Health Sciences.; 5(5):75-82
View at Publisher | View at Google Scholar - Chaudhary R, Thapa SK, Rana JC, Shah PK. (2017 )., Surgical site infections and antimicrobial resistance pattern. Journal of Nepal Health Research Council. 15(2): 120-123.
View at Publisher | View at Google Scholar - Yimer RM, Alemu MK. (2022),.Bacterial Contamination Level of Indoor Air and Surface of Equipment in the Operation Room in Dil-Chora Referral Hospital, Dire Dawa, Eastern Ethiopia. Infect Drug Resist.; 15:5085-5097
View at Publisher | View at Google Scholar

 Clinic
Clinic
