Research Article | DOI: https://doi.org/10.31579/2835-8325/064
A Study of Ewsr1 Gene Rearrangement by Fluorescence In-Situ Hybridization in Ewing’s Sarcoma Patients
Cytogenetics Laboratory, Cancer Biology Department, The Gujarat Cancer and Research Institute, Asarwa, Ahmedabad-380016, Gujarat, India
*Corresponding Author: Pina J. Trivedi, Cytogenetics Laboratory, Cancer Biology Department, The Gujarat Cancer and Research Institute, Asarwa, Ahmedabad-380016, Gujarat, India.
Citation: Dharmesh M. Patel, Dhrumil Parmar, Pina J. Trivedi, Mahnaz M. Kazi, et al, (2024), A Study of Ewsr1 Gene Rearrangement by Fluorescence In-Situ Hybridization in Ewing’s Sarcoma Patients, Clinical Research and Clinical Reports, 4(2); DOI:10.31579/2835-8325/064
Copyright: © 2024, Pina J. Trivedi. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Received: 07 February 2024 | Accepted: 23 February 2024 | Published: 20 March 2024
Keywords: laser; patients
Abstract
Introduction: Ewing's sarcoma is a rare form of cancer affecting bones or the adjacent soft tissues, primarily seen in individuals aged between 10 to 30 years. As the second most common malignant bone tumor in this age group, it constitutes around 6% to 10% of all malignant bone tumors. Notably, its identification is linked to the EWSR1 gene translocation, making Fluorescence In-Situ Hybridization (FISH) analysis pivotal for accurate diagnosis. Objective of the present study to scrutinize the correlation between the EWSR1 gene translocation and the clinicopathological characteristics in patients diagnosed with Ewing's sarcoma. This research aims to investigate EWSR1 gene translocation through the Fluorescence In-Situ Hybridization (FISH) technique. This approach holds significance as it involves mastering FISH techniques, assessing the link between EWSR1 gene translocation, and exploring how it relates to various clinicopathological aspects in patients with Ewing's Sarcoma.
Method: Histopathologically confirmed patients with Ewing’s sarcoma were included. EWSR1 gene rearrangement was detected in total 35 patients with Ewing’s sarcoma tissue using Fluorescence In-Situ Hybridization. FISH was performed using EWSR1 Dual Color Break Apart probe kit.
Result: 35 patients were enrolled in the study, among them 60% (21/35) of the patients were male and 40% (14/35) of the patients were female and the median age was 16 years. EWSR1 gene rearrangement was positive in 71% (25/35) of the patients while 29% (10/35) of the patients were negative. EWSR1 gene rearrangement was significantly associated with site and nearly significant with lymph node involvement but not with age, gender, size, lung metastasis, and not significant association with histopathological parameters.
Summary and Conclusion: This study illustrates the importance of FISH as an ancillary diagnostic tool in the diagnosis of EWSR1 rearranged neoplasms. EWSR1 gene rearrangement is significantly higher in soft tissue followed by bone and parenchymal organs. Present study shows positive for EWSR1 rearrangement by FISH technique in 2 (9%) CD99 negative patients, 6 were weak positive patients and 3 (14%) were weakly positive for FLI1 by Immunihistochemistry (IHC) this signifies the role of molecular studies in cases difficult to diagnose on routine histopathology and IHC.
Introduction
Ewing’s Sarcoma can occur at any age but, the peak incidence occurs in the second decade of life, and males suffer from ES about one and half times more often than females. (Javed MU et. al.; 2009) Generally, ES can occur at any part of the body but, most commonly affects bone particularly pelvis, femur or axial skeleton and, it can also involve soft tissues in upto 30% of patients. (Applebaum MA. et. al.; 2011), (Ordonez JL. et. al.; 2009), (Kauer M. et. al.; 2009) However, the diagnosis is reached in most cases only with the help of IHC. On the other hand, some cases require molecular studies for the confirmation of the diagnosis. (Gambheri G. et. al.; 2011) Gene rearrangement resulting in a translocation are a defining diagnostic feature in many hematopoietic and solid tumors. Detection of specific gene rearrangement by Fluorescence In-Situ Hybridization (FISH) is commonly used in practice of pathology, in addition to immunohistochemistry, to aid in the diagnosis of more difficult cases. Moreover, FISH method is found to be more sensitive and specific compared to Reverse Transcriptase–Polymerase Chain Reaction (RT-PCR) on Formalin Fixed Paraffin Embedded (FFPE) tissue section and consists of 91% of sensitivity and 100% specificity. Therefore, it is easier to interpret the results. (Bridge RS. Et. al.; 2006) Ewing sarcoma breakpoint region 1 (EWSR1) gene is located at 22q12 translocation involving the EWSR1 gene was first described in and first to molecularly defined Ewing’s Sarcoma. (Delattre O. et. al.; 1994) The majority of Ewing’s Sarcoma family of tumors is defined by a translocation resulting in the fusion of the EWSR1 gene and the gene of the E26 transcription-specific (ETS) family of transcriptors factor such as FLI1, ERG, ETV1, E1AF and FEV. hotmail.com (Cantile M. et. al.; 2013) The aim of this research is to study EWSR1 gene translocation by Fluorescence In-Situ Hybridization (FISH) technique which is significant because it involves learning FISH technique, evaluating the association of EWSR1 gene translocation using FISH technique and correlating EWSR1 gene translocation with various clinicopathological characteristics of Ewing’s Sarcoma patients.
Materials and Method
Patient Details: This retrospective study, sanctioned by the institutional scientific and ethics committee, encompassed 35 cases of Ewing's Sarcoma patients diagnosed morphologically and immunohistochemically at The Gujarat Cancer & Research Institute (GCRI). All cases involved excision specimens, with exclusions made for those with inconclusive diagnosis of ES and specimens lacking proper formalin fixation resulting in cellular detail loss. Macroscopic examination of the specimens adhered to established guidelines, followed by staining with Hematoxylin and Eosin (H&E) and a panel of immunohistochemical stains including vimentin, MIC-2 (CD99), TdT, synaptophysin, FLI1, and desmin immunostain. Pertinent clinicopathological data such as age, gender, tumor size, histopathological type, stage, and metastasis were retrieved from the maintained case files in the Medical Record Department of the institute. Disease status evaluation was conducted through clinical examination, radiological investigations, and biochemical assessments. (Table 1)
| CHARACTERS | N(%) |
| Age (Median: 16 years, Range: 1-60) | |
| ≤16 years | 18(51) |
| >16 years | 17(49) |
Gender | |
| Male | 21(60) |
| Female | 14(40) |
Tumor size | |
| ≤3cm | 25(86) |
| 3.1-8cm | 02(07) |
| >8cm | 02(07) |
Site | |
| Bone | 21(60) |
| Soft tissue | 10(29) |
| Parenchymal organs | 04(11) |
Lymph node | |
| Negative | 25(71) |
| Positive | 10(29) |
Lung metastasis | |
| Negative | 33(94) |
| Positive | 02(06) |
Stage | |
| I | 18(56) |
| IV | 14(44) |
FLI1 | |
| Negative | 02(09) |
| Weak positive | 03(14) |
| Positive | 17(77) |
CD99 | |
| Weak positive | 09(26) |
| Positive | 26(74) |
| Desminimmunostain | |
| Negative | 35(100) |
Vimentin | |
| Negative | 03(21) |
| Weak positive | 04(29) |
| Positive | 07(50) |
Synaptophysin | |
| Negative | 11(73) |
| Weak positive | 02(13) |
| Positive | 02(13) |
TdT | |
| Negative | 35(100) |
Table 1: Clinicalcharacteristic of the EWSR1 gene rearrangement patients
Fluorescence In-Situ Hybridization (FISH) technique: FISH analysis was conducted on paraffin blocks using the LSI EWSR1 Dual Color Break-Apart probe (Zytovision). Tumor tissue sections, 4 µm thick, were utilized for the FISH studies on coated slides. Post- deparaffination, a series of washes involving xylene, alcohol, and TDW were administered. Slides underwent Pre-treatment solution treatment at 95°C for 25 minutes, followed by pepsin treatment at room temperature for 20 minutes. For the hybridization process, 1 µl of the probe was applied to the target area, covered with a coverslip, sealed with rubber cement, and then incubated at 37°C overnight. The subsequent day, the coverslip was removed, and the slide underwent post-hybridization washes, with 1 drop of DAPI counterstain applied before placing the coverslip. Observation of the slide was carried out using an Epi- fluorescence microscope, and scoring and capture were performed using Isis software.
Statistical analysis
The statistical analysis in this study was conducted using SPSS statistical software, version 26, (developed by SPSS Inc., U.S.). Key descriptive statistics, including mean values, standard error of the mean (SE), and median values, were computed to summarize the data. To examine the relationship and significance between two variables, Pearson's Chi-square (χ2) test and Pearson's correlation coefficient (r) were employed as statistical tools. In accordance with common statistical conventions, P values less than or equal to 0.05 were deemed indicative of statistically significant findings.
Result
Out of total 35 (100%) patients, 10 (29%) patients showed 2F signal pattern. Hence, these patients were considered negative for EWSR1 rearrangement. While 25 (71%) patients showed 1O1G1F (OGF) signal pattern considered positive for EWSR1 rearrangement (Figure 2).
Figure 2 Representative imagesof FISH resultsfor EWSR1 gene rearrangement
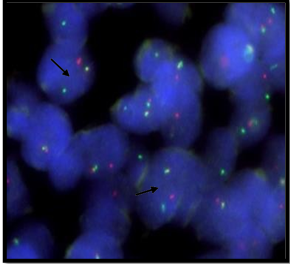
Figure 2A: Positive result for EWSR1 gene rearrangement
Image showsOGY pattern describing 1O1G1F signal showspositive result for EWSR1 gene rearrangement.
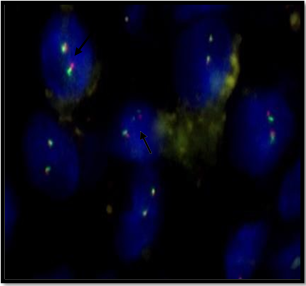
Figure 2B: Negative result for EWSR1 gene rearrangement
Image shows YY pattern describing 2F signal shows negative result for EWSR1 gene rearrangement.
Among the total enrolled patients, 71% (25/35) patients were positive for EWSR rearrangement, while 29% (10/35) were negative. Out of total patients 60% (21/35) were male, among that 67% (14/21) exhibiting positive EWSR1 rearrangement and 33% (7/21) showing negative results. Additionally, among the 14 female patients, 79% (11/14) were positive for EWSR1 rearrangement, while 21% (3/14) were negative. χ2=0.583, r = -0.129 and p= 0.460. (Table 2)

Table 2: correlation of EWSR1 gene with clinicopathological parameters
From the entire patient, 51% (18/35) patients were 16 years or younger, among that 72% (13/18) were positive for EWSR1 rearrangement and 28% (5/18) were for negative results. Furthermore, within the group, 49% (17/35) were female, of which 71% (12/17) tested positive and 29% (5/17) were negative for EWSR1 rearrangement. χ2=0.011, r= 0.018 and p=0.918. (Table:2)
The study encompassed three primary sites: Bone (60%), Soft tissue (29%), and Parenchymal organs (11%). Among the patients with bone involvement, 76% (16/21) tested positive, while 24% (5/21) tested negative for rearrangement. (Figure 1)
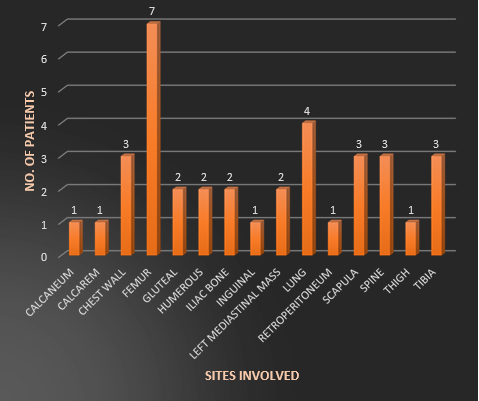
Figure 1: different sites involved in Ewing’s Sarcoma
In the case of soft tissue, 80% (8/10) of patients showed a positive result, and 20% (2/10) displayed a negative outcome. Furthermore, among patients with parenchymal organ involvement, 25% (1/4) tested positive, while 75% (3/4) tested negative for EWSR1 rearrangement. Notably, the study revealed a highly significant association between the specific sites and the EWSR1 gene. χ2=7.560, r= - 0.421, p=0.012. (Table 2, Figure 3)
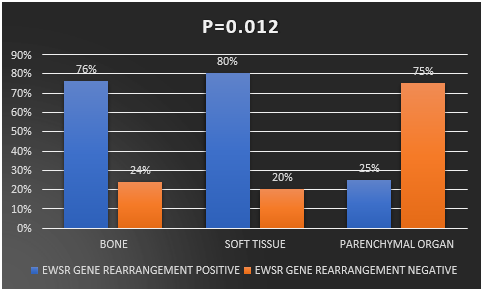
Figure 3: Correlation between EWSR1gene rearrangement with sites
Lymph node involvement was detected in 29% (10/35) of the patients, with 50% (5/10) testing positive and 50% (5/10) testing negative for EWSR1 gene rearrangement. Conversely, 71% (25/35) of the patients did not show lymph node involvement, with 80% (20/25) testing positive and 20% (5/25) testing negative for EWSR1 gene rearrangement. This study indicated a near significance between lymph node involvement and EWSR1 gene rearrangement. χ2= 3.150, r= -0.300, p= 0.080. (Table 2, Figure 4)
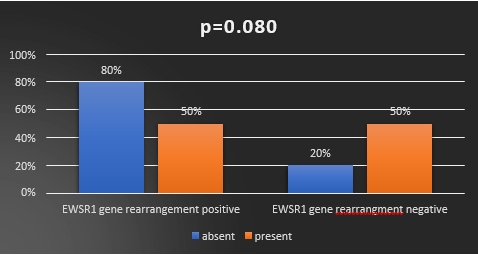
Figure 4: Correlationbetween EWSR1 gene rearrangement with lymph node metastasis
Ewing's sarcoma is a highly aggressive cancer comprised of undifferentiated round cells. Its
Discussion
Ewing's sarcoma is a highly aggressive cancer comprised of undifferentiated round cells. Its
exact origin is uncertain. This cancer typically affects individuals between the ages of 10 and 30, and it is known for its rapid growth and tendency to spread to other parts of the body. (Lin PP. et. al.; 2011) Ewing's sarcoma exhibits a peak incidence during the second decade of life,
with a notable predilection for males, affecting them at a rate approximately one and a half times higher than females. (Riggi N. et. al.; 2007) Ewing's sarcoma typically presents a predilection for diverse anatomical sites within
the body. While it may manifest in virtually any location, it most frequently afflicts the skeletal system, with a notable predilection for the femur, pelvis, scapula, and the axial skeleton. Nevertheless, this malignancy also demonstrates an ability to infiltrate soft tissues and parenchymal organs in affected patients,
underscoring its capacity for anatomical versatility. (Applebaum MA. et. al.; 2011)
Ewing Sarcoma, denoted as ESFT (Ewing Sarcoma Family of Tumors), is distinguished by non- random chromosomal translocations, which culminate in the formation of fusion genes
positioned at the C-terminus of the transcribed region. These fusion proteins harbour a distinctive feature, namely the presence of RGG repeats that facilitate their binding to RNA molecules, thereby actively engaging in transcriptional processing and RNA splicing. The N-terminus of the fusion protein originates from the ETS family of transcription factors, with members such as FLI1, ERG, FEV, and others playing a pivotal role in this context. Notably, the t (11;22) (q24; q12) translocation is a hallmark event, observed in 85-90% of Ewing Sarcoma tumors, effectively generating the fusion protein. The remaining 10- 15% of cases, on the other hand, entail alternative translocations such as t (7;22) (p22; q12) and other less common chromosomal rearrangements. (Riggi N. et. al.; 2007)
The identification of EWSR1 gene rearrangements can be accomplished through FISH utilizing
Break-Apart probes on FFPE tissue specimens. FISH stands out as a prominent and extensively employed technique for molecularly diagnosing ESFT due to its ability to be executed with minimal tissue samples and its compatibility with FFPE materials. This method is increasingly gaining prevalence in numerous medical centers as a pivotal tool for confirming the molecular diagnosis of ESFT. (Murthy SS. et. al.; 2021) FISH offers two distinct approaches for analysis, namely the fusion technique and the break-apart technique, both of which consistently yield congruent results. The choice to employ the FISH break-apart strategy is typically made when multiple translocation partners are involved in the genetic rearrangement under investigation. This strategy proves to be a cost-effective alternative, marked by its capacity to generate brighter and larger signal patterns, rendering interpretation of the results a more straightforward and efficient process. (Bridge RS. et. al.; 2006)
In the present investigation, a rearrangement of the EWSR1 gene was detected in 71% (25/35) of the patients, consistent with the findings of Murthy S. et al. (2021) who reported a 79% incidence, and Bashir M. et al. (2020) who identified an 88% occurrence of the EWSR gene rearrangement. Various published studies have documented the presence of EWSR1 gene rearranged tumors in the diagnosis of ESFT, with reported rates of 92% (144/156) (Gambheri G. et. al., 2011), 82% (89/109) (Noujaim J. et al.; 2017), 91% in a series with no false positives (Bridge RS. et al.; 2006), and 83% (15/18) of cases (Qian X. et al.; 2005). Indian studies have also reported EWSR1 positive outcomes via FISH in a limited sample size, with rates of 91% (10/11) (Jambhekar NA et al.; 2006) and 92% (12/13) of cases (Rekhi B. et al.; 2014).
The high incidence of EWSR1 gene rearrangement was found in soft tissue for about 80% of the patients. The study found significant correlation between sites with EWSR1 gene rearrangement. (p=0.012) Similar studies was done by Bashir M. et. al. study and they found 51% had bone involvement. (Bashir MR. et. al.; 2020)
To conclude, FISH emerges as a robust and dependable adjunctive technique in the diagnostic armamentarium for identifying Ewing's sarcoma and pinpointing the translocation partner genes involved. This investigation underscores the pivotal role of FISH as a supplementary diagnostic tool in the evaluation of neoplasms featuring EWSR1 gene rearrangements. Notably, the prevalence of EWSR1 gene rearrangements is notably higher in soft tissue tumors, followed by occurrences in bone and parenchymal organs. Within our study cohort of 25 patients, FISH analysis identified 6 patients with weak CD99 positivity, while 2 patients (comprising 9%) exhibited a lack of CD99 expression, and 3 patients (14%) showed only weak FLI1 positivity by IHC. This underscores the invaluable contribution of molecular studies in cases that present diagnostic challenges through routine histopathological and IHC analysis.
References
- Lin PP, Wang Y, Lozano G. (2011). Mesenchymal Stem Cells and the Origin of Ewing’s Sarcoma. Sarcoma, 276463.
View at Publisher | View at Google Scholar - Jawad MU, Cheung MC, Min ES, Schneiderbauer MM, Koniaris LG, Scully SP.l (2009). Ewing sarcoma demonstrates racial disparities in incidence-related and sex-related differences in outcome: an analysis of 1631 cases from the SEER database, 1973- 2005. Cancer, 115:3526-3536.
View at Publisher | View at Google Scholar - Applebaum MA, Worch J, Matthay KK, Goldsby R, Neuhaus J, West DC, et al. (2011). Clinical features and outcomes in patients with extraskeletal Ewing sarcoma. Cancer, 117:3027-3032.
View at Publisher | View at Google Scholar - Ordonez JL, Osuna D, Herrero D, de Alava E, Madoz-Gurpide J. (2009). Advances in Ewing’s sarcoma research: where are we now and what lies ahead? Cancer Res, 69:7140-7150.
View at Publisher | View at Google Scholar - Kauer M, Ban J, Kofler R, Walker B, Davis S, Meltzer P, et al. (2009). A molecular function map of Ewing’s sarcoma. PLoS ONE, 4:5415.
View at Publisher | View at Google Scholar - Gambheri G, Cocchi S, Benini S, et al. (2011). Molecular diagnosis in Ewing family tumors: the Rizzoli experience—222 consecutive cases in four years. J Mol Diagn, 13:313-324.
View at Publisher | View at Google Scholar - Bridge RS, Rajaram V, Dehner LP, Pfeifer JD, Perry A. (2006). Molecular diagnosis of Ewing sarcoma/primitive neuroectodermal tumor in routinely processed tissue: a comparison of two FISH strategies and RT-PCR in malignant round cell tumors. Mod Pathol, 19:1-8.
View at Publisher | View at Google Scholar - Delattre O, Zucman J, Melot T, Garau XS, Zucker JM, Lenoir GM, et al. (1994). The Ewing Family of Tumors--a Subgroup of Small-Round-Cell Tumors Defined by Specific Chimeric Transcripts.” The New England Journal of Medicine, 331: 294-299.
View at Publisher | View at Google Scholar - Tony LNg, O’Sullivan, Pallen CJ, Malcolm Hayes, Clarkson pw, Mark Winstanley, et al. (2007). Ewing Sarcoma with Novel Translocation t (2;16) Producing an In-Frame Fusion of FUS and FEV. The Journal of molecular diagnostics: JMD, 9: 459-463.
View at Publisher | View at Google Scholar - Cantile M, Marra L, Franco R, Ascierto P, Liguori G, Chiara AD, et al. (2013). Molecular detection and targeting of EWSR1 fusion transcripts in soft tissue tumors. Medical Oncology (Northwood, London, England), 30: 412.
View at Publisher | View at Google Scholar - Riggi N, Cironi L, Suvà ML, Stamenkovic I. Sarcomas. (2007). genetics, signalling, and cellular origins. Part 1: The fellowship of TET. The Journal of pathology, 213(1):4-20.
View at Publisher | View at Google Scholar - Murthy SS, Gundimeda SD, Challa S, Manjula V, Fonseca D, Rao VB, Rajappa SJ, Raju KV, Rao TS. (2021). FISH for EWSR1 in Ewing's sarcoma family of tumors: Experience from a tertiary care cancer center. Indian Journal of Pathology and Microbiology, 1;64(1):96.
View at Publisher | View at Google Scholar - Bashir MR, Pervez S, Hashmi AA, Irfan M, Hashmi AA. (20020). Frequency of translocation t (11; 22) (q24; q12) using fluorescence in situ hybridization (FISH) in histologically and immunohistochemically diagnosed cases of Ewing’s sarcoma. Cureus, 20:12(8).
View at Publisher | View at Google Scholar - Noujaim J, Jones RL, Swansbury J, Gonzalez D, Benson C, Judson I, Fisher C, Thway K. (2017). The spectrum of EWSR1-rearranged neoplasms at a tertiary sarcoma centre; assessing 772 tumour specimens and the value of current ancillary molecular diagnostic modalities. British Journal of Cancer, 116(5):669-678.
View at Publisher | View at Google Scholar - Qian X, Jin L, Shearer BM, Ketterling RP, Jalal SM, Lloyd RV. (2005). Molecular diagnosis of Ewing's sarcoma/primitive neuroectodermal tumor in formalin-fixed paraffin- embedded tissues by RT-PCR and fluorescence in situ hybridization. Diagnostic Molecular Pathology, 14(1):23-28.
View at Publisher | View at Google Scholar - Jambhekar NA, Bagwan IN, Ghule P, Shet TM, Chinoy RF, Agarwal S, Joshi R, Amare Kadam PS. (2006). Comparative analysis of routine histology, immunohistochemistry, reverse transcriptase polymerase chain reaction, and fluorescence in situ hybridization in diagnosis of Ewing family of tumors. Archives of pathology & laboratory medicine. 130(12):1813-1818.
View at Publisher | View at Google Scholar - Rekhi B, Vogel U, Basak R, Desai SB, Jambhekar NA. (2014). Clinicopathological and molecular spectrum of Ewing sarcomas/PNETs, including validation of EWSR1 rearrangement by conventional and array FISH technique in certain cases. Pathology & Oncology Research, 20:503-516.
View at Publisher | View at Google Scholar

 Clinic
Clinic
