Review Article | DOI: https://doi.org/10.31579/2835-785X/108
Pharmacogenetics: Role of Pharmacogenetics in Treating Various Diseases
1 Research student, Department of Pharmaceutics, RBVRR Women’s College of Pharmacy, Barkatpura, Osmania University, Hyderabad, Telangana, India.
2 Professor, Department of Pharmaceutics, RBVRR Women’s College of Pharmacy, Barkatpura, Osmania University, Hyderabad, Telangana, India.
*Corresponding Author: Dr. A Krishna Sailaja, Professor, Department of Pharmaceutics, RBVRR Women’s College of Pharmacy, Barkatpura, Osmania University, Hyderabad, Telangana, India.
Citation: Sadiyah Sultana, and Dr A Krishna Sailaja, (2025), Pharmacogenetics: Role of Pharmacogenetics in Treating Various Diseases, International Journal of Clinical Research and Reports. 4(5); DOI: 10.31579/2835-785X/108.
Copyright: © 2025, Dr. A Krishna Sailaja. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Received: 22 September 2025 | Accepted: 22 October 2025 | Published: 14 November 2025
Keywords: Pharmacogenetics; single nucleotide polymorphism; pharmacogenomics; personalized medicine; genetic variation; genetic disorders
Abstract
Pharmacogenetics is essential in managing serious diseases by customizing drug treatments according to a person’s genetic profile. This field enhances the effectiveness of medications while reducing the risk of side effects in illnesses such as cancer, diabetes, autoimmune and mental health issues. By analyzing genetic differences that influence how drugs are processed and how the body responds, pharmacogenetics supports the development of individualized treatment strategies. This personalized approach leads to better health outcomes, fewer hospital admissions and decreased medical expenses. For instance, in cancer treatment, it assists in choosing the most suitable targeted therapies, it helps identify the safest and most effective drugs. Ultimately, pharmacogenetics advances precision medicine by improving the safety and efficiency of treatments for life-threatening diseases.
Introduction
Pharmacogenetics: The term pharmacogenetics is about the study of how differences in a genetic sequence affect individual response to medication [1].
Friedrich Vogel coined the term “Pharmacogenetics” in the year 1959.
The term “PHARMACOGENOMICS” is also used due to their overlapping scope in studying genetic influence on drug response [2].
"Pharmacogenetics typically focuses on the impact of individual genetic variants, whereas pharmacogenomics encompasses a wider perspective, examining how multiple genetic variations across the entire genome influence a person's response to medications.
Many genetic and non-genetic factors which includes individuals age, concomitant therapy affect the response to therapies where genetics influences can be primary reason for 95% of the differences observed in treatment responses.[3]
Need of Pharmacogenetics: [4,5]
In not more than 60%of patients prescribed medicines are effective and a significant number of patients also develop major adverse effects. The goal is to utilize genetic differences to select the most effective medication, an optimal dosage, and an appropriate treatment duration in order to maximize therapeutic benefits while minimizing the risk of adverse effects for an individual patient. It helps in predicting response to drugs addressing both efficacy and safety issues
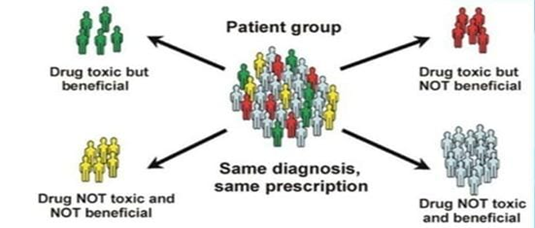
Figure 1: Significance of Pharmacogenetics.
Genetic Basis of Variable Drug Response: [6]
The pharmacokinetics of drugs, along with their therapeutic and toxic effects (pharmacodynamics), are governed by intricate biological processes involving proteins encoded by various genes that affect drug transport, metabolism and mechanisms of action.
Table 1: Examples of Differences in Gene Variants That Affect Drug Response in Humans:[6]
Drug | Variable clinical effect | Genes with associated variants | Possible mechanism |
Azathioprine | Elevated haematopoietic toxicity | TPMT | Decrease in function TPMT gene expression |
Irinotecan | Elevated haematopoietic toxicity | UGT1A1 | Decrease in expression |
Fluorouracil | High toxicity | DPD | Suppression in enzymatic activity due to mutation |
Omeprazole | Peptic ulcer response | CYP2C19 | Low activity gene variant |
β-blockers | Decreased activity | CYP2D6 | Gene duplication |
Warfarin | Increased anticoagulant effects | CYP2C9 | Coding region variants causing reduced S-warfarin clearance |
Abacavir | Immunologic reactions | HLA variants | Altered immunologic responses |
Personalized Medicine
Personalized medicine is also often addressed as precision medicine. Personalized medicine is commonly known as P4 medicine, which stands for predictive, preventive, personalized, and participatory healthcare [7].
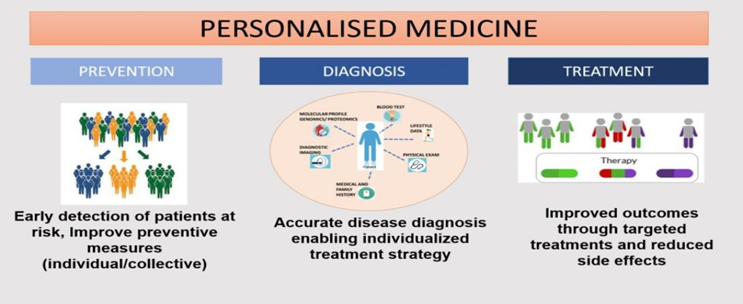
Figure 2: Various steps in personalized medicine.
Advantages:[8]
1. Prediction of susceptibility to disease and accurate diagnosis of disease.
2. Helps selecting medications that are more effective while avoiding those likely to cause known adverse effects.
3.Tailoring strategies for preventing diseases to individual needs.
4.Enhancing patient compliance by matching the appropriate medication to the appropriate individual.
5. Safe and effective treatment is achieved.
6. Significant reduction in healthcare costs.
7.Useful for early prevention and the identification of previously undetected health conditions.
Categories of Patient
Patients are mainly classified depending on genetic polymorphism.[9]
- Genetic Polymorphism: Genetic polymorphism is the variation in sequence of DNA between different group and population. Genetic variations related to a drug's pharmacokinetics and pharmacodynamics can lead to toxic effects, altered efficacy, or lack of therapeutic response. These genetic differences often serve as biomarkers for diagnosing diseases and guiding therapeutic decisions.
- Single nucleotide polymorphism (SNP’s): Variation in a single allele of a gene involved in various metabolic functions, including the metabolism of enzymes.
The different alleles which makeup a gene indicates the activity of enzyme and determines to what extent the enzyme is active.SNP’s which do not alter the encoded amino acids are called synonymous SNPs whereas the SNPs which change the encoded amino acids are non-synonymous SNPs.
Phase 1 Enzyme Polymorphism:[10]
- CYP2D6: This enzyme is responsible for metabolism of around 30% of drugs.
Table 2: Impact of variations in CYP2D6 on Metabolism.
Type of metabolizers | Type of allele | Name of enzyme |
Slow metabolizers | 2 defectives | CYP2D6*4/*4 CYP2D6*4/*5 |
Intermediate metabolizes | 1 defective, 1 wild type | CYP2D6*4/*5 & CYP2D6*1/*3 |
Normal metabolizer | 2 wild types | CYP2D6*1/*3 |
Fast metabolizers | 1 wild type, 1 amplified | CYP2D6*1/*2& CYP2D6*1A/*5 |
Ultra-rapid metabolizers | 2 or more amplified | CYP2D6*2/*3 |
- CYP1A2:
It plays a role in metabolizing approximately 5% of drugs available on the market, such as fluvoxamine, clozapine, olanzapine and theophylline. About 15% of the Japanese population, and 5% of both Chinese and Australians, are identified as poor metabolizers of the CYP1A2 enzyme.
- CYP2C9:
CYP2C9 possesses over 30 known allelic variants, with two of them being the most common which include CYP2C9*2 /*3. These variants lead to decreased CYP2C9 enzyme activity, which is observed in approximately 35% of the Caucasian population. It plays a key role in the metabolism of warfarin, a blood thinner with a narrow therapeutic index.
- CYP2C19:
It is a drug-metabolizing enzyme with high genetic variability, having over 30 different known variants. It is involved in metabolism of clopidogrel and several antidepressants.
- CYP3A4:
It is the most prevalent CYP450 enzyme in the liver and is responsible for metabolizing more than 50% of commonly prescribed drugs, with over 20 allelic variants identified [11].
Phase 2 Enzyme Polymorphism:
- N-acetyl transferase: It was identified as a polymorphic enzyme through phenotypic observations of fast and slow acetylators of antituberculotic drug, Isoniazid.[12] NAT activity is coded by two gene which are NAT1 and NAT2. The fast and slow acetylators phenotype was found to be associated with NAT2 gene. The slow acetylator trait occurs in approximately half of Caucasians, around 90% of individuals of Arab descent, and about 10% of the Japanese population.[13]
- Thiopurine methyltransferase: Metabolizes drugs with thiopurine used in treatment of leukemia's and autoimmune disorders.
A patient with a genetic mutation may be unable to produce this enzyme, which can cause thiopurine to accumulate in the body, leading to potential complications. At least 28 allelic TPMT variants have been discovered, with the majority of null phenotypes linked to the TPMT2, TPMT3A, and TPMT*3B alleles, which lead to non-synonymous mutations.
- UGT1A1 enzyme: Metabolizes Irinotecan used in chemotherapy of colorectal cancer. Patient with genetic variation may observe increase buildup of the drug [14].
Doctors can test for genetic variation before treatment and customize the dose of the drug to be given.
Role of pharmacogenetics in treating various diseases
1. Cystic Fibrosis
Cystic fibrosis is a disorder which is genetically inherited, it damages the mucous lining of digestive system and lungs. One of the most egregious examples of how diagnosis and treatment were improved using Pharmacogenetics is cystic fibrosis (CF). Cystic fibrosis transmembrane conductance regulator (CFTR) gene mutation results in development of cystic fibrosis. Drugs that can (at least partially) restore CFTR channel function on a molecular basis have been developed and approved Ex: Ivacaftor and lumacaftor [15]. Other medications like NSAIDS & Corticosteroids are used to reduce the inflammation occurring in cystic fibrosis.
However, these medications contribute to drug therapy problems in patients with polymorphism in CYP2D6 and CYP2C19.Pharmacogenetics testing can help in reducing drug therapy problems in patient suffering with cystic fibrosis and guide in pharmacotherapy. Patients with mutation in G551D are treated with Ivacaftor while F508 mutated gene is treated using elexacaftor and tezacaftor. This indicates that pharmacogenetic testing help in appropriate selection of the drug [16].
2. Monogenic Diabetes
Monogenic diabetes, which results from a mutation in a single gene, serves as a notable example of the effective use of personalized medicine.Most cases of Maturity Onset Diabetes of the Young (MODY) are due to mutations in the HNF1 and HNF4 genes, corresponding to MODY types 3 and 1, respectively[17].Patients with MODY types 1 and 3 are typically diagnosed before the age of 25 and usually do not show any indications of insulin resistance.In few cases these patients are diagnosed incorrectly with Type 1 diabetes, for which insulin therapy is given. However, with an accurate diagnosis, these patients may begin treatment with sulfonylureas [18].
3. Attention Deficit Hyperactivity Disorder (ADHD)
ADHD is a condition marked by ongoing and developmentally unsuitable patterns of inattention, hyperactivity, and impulsive behavior. The commonly used medications for treatment of ADHD are methylphenidate, atomoxetine, clonidine, guanfaine and amphetamine salts. Atomoxetine is metabolised by CYP4502D6 whereas methylphenidate is metabolised by carboxylesterase 1 [19]. Patients who are poor metabolizers, administration of atomoxetine should up to 10 folds higher exposure than the normal metabolizers at same dose.
Similarly, patients upon administration of methylphenidate rs71647871 variant show 2-fold higher exposure than the normal individuals [20]. Study of these genes helps in administration of appropriate drugs with appropriate dose in order to treat ADHD.
4. Inflammatory Bowel Disease (IBD) [21]
Inflammatory bowel disease includes Crohn's disease and ulcerative colitis, both marked by long-lasting inflammation of the intestines. PPAR-γ plays a part in controlling this intestinal inflammation. Mesalazine is considered the primary treatment option in patients clinically diagnosed with remission in ulcerative colitis. It works directly on the colonic lining to decrease inflammation by activating PPAR-γ through multiple anti-inflammatory mechanisms [21].
A genetic study revealed that IBD patients with SNO rs 2647087 within HLADQA1 upon administration of thiopurines resulted in increased risk of pancreatitis.
Another study revealed that genetic polymorphism of enzyme glutathione s transferase results in decreased sensitivity to Azathiopurine.
In Asian patients NUDT15 was found to be a genetic marker for thiopurine tolerance. And polymorphism of this gene resulted in myelosuppression and hair fall. This allows in reducing the dosing frequency of thiopurines for Asian patient suffering with IBD. Individuals lacking sufficient TMPT enzyme activity face a threefold increased risk of gastrointestinal side effects [22].
5. Rheumatoid Arthritis [23,24]
Rheumatoid arthritis (RA) is an autoimmune disorder impacting approximately 1% of people, characterized by persistent inflammation of the synovial joints that can result in pain and loss of joint function. Methotrexate is the most frequently used disease-modifying antirheumatic drug (DMARD) for treating rheumatoid arthritis. Changes in the MTHFR gene can affect how methotrexate works, potentially causing unforeseen side effects and increased toxicity. The common variants of the MTHFR gene include 677 C>T and 1298 A>C.
Administration of methotrexate with 677 C>T polymorphism impairment in enzyme activity results in increased toxicity of methotrexate and hyper homocysteinee Mia. Similarly in patients with 1298A>Cpolymorphism also results in hyper homocysteine Mia.
Patients suffering rheumatoid arthritis with elevated TYMS gene expression requires higher dose of methotrexate.
NAT gene responsible for metabolism of sulfasalazine. NAT 2*4 polymorphism showed toxicity in slow acetylators and increased efficacy in rapid acetylators. Hence based on genetic data drug dose can be calculated.
6. Melanoma: [25,26]
The BRAF gene in humans produces a protein known as Braf, which is essential for signaling pathways that regulate cell growth and is often found to be mutated in various cancers. In 2011, vemurafenib, a drug that inhibits the Braf protein, along with a diagnostic test for the BRAF V600E mutation, received approval for treating advanced-stage melanoma. Vemurafenib is effective particularly in treating patients whose tumors have been identified with the V600E mutation in the BRAF gene.
7. Breast Cancer: [27,28]
Trastuzumab represents one of the first and most well-known examples of personalized medicine in the treatment of breast cancer.
Patients suffering from breast cancer, around 30% of the mare observed to have over expression of a protein known as HER2 that does not respond to conventional treatments. Trastuzumab received approval in 1998 for use in patients with HER2-positive tumors, and subsequent studies in 2005 demonstrated that, when combined with chemotherapy, it reduced cancer recurrence by 52%. However, the efficacy of transtuzumab in HER2 positive patients is dependent on antibody dependent cell mediated cytotoxicity (ADCC) governed by FCGR3A gene.
Antiestrogen treatments, including selective estrogen receptor modulators and aromatase inhibitors, are advised for ER-positive tumors, but not for those that are ER-negative. Tamoxifen is the commonly used SERM, which is used to treat of malignant breast cancer. It was observed the CYP2D6 gene responsible for tamoxifen bioactivation has several genetic polymorphisms which results in poor metabolizers and extensive metabolizers could affect its enzyme activity and thereby results in ineffective therapeutic activity.
Challenges: [29]
1. Offering scientific support for enhancing patient care through pharmacogenomic testing
2. Development of new genomic technologies.
3. Healthcare Professionals education.
4. Patient acceptance.
5. Ethical issues.
6. Costly.
7. Regulatory issues in gene testing.
Conclusion
Pharmacogenetics plays a crucial role in ensuring the safety and effectiveness of medications. It examines how variations in individual genes or combinations of genes influence a person’s response to drugs. Personalized medicine utilizes genetic information to address individual differences among patients, leading to better diagnostic accuracy, more effective treatments, improved disease prevention, and increased cost-efficiency. Today, pharmacogenomics is a key component of drug development, with many pharmaceutical companies leveraging genetic data to discover new drug targets and to identify specific patient subgroups likely to benefit from experimental therapies. By understanding how a person's genetic profile affects their response to medications, pharmacogenomics supports more informed and precise clinical decision making.
References
- Jeffrey R. Bishop, in Handbook of Clinical Neurology, Neurogenetics 2018.
View at Publisher | View at Google Scholar - Pooja D Gupta et al., (2015), Pharmacogenetics, Pharmacogenomics and Ayurgenomics for Personalized Medicine: A Paradigm Shift Indian Journal of Pharmaceutical Science;77(2):135-141
View at Publisher | View at Google Scholar - Oates JT et al., Pharmacogenetics: An Important Part of Drug Development with a Focus on Its Application. International journal of Biomedical Investigation.1(2):111.
View at Publisher | View at Google Scholar - Mc Carthy AD., et al., Pharmacogenetics in drug development. Philosophical transactions of the Royal Society of London B Biological Sciences 2005; 360(1460):1579-1588
View at Publisher | View at Google Scholar - Roses AD. (2000), Pharmacogenetics and the practice of medicine. Nature Reviews; 405:(6788):857-865.
View at Publisher | View at Google Scholar - Enrico Mini and Stefania Nobili et al., (2009), Pharmacogenetics: implementing personalized medicine Clinical Cases in Mineral and Bone Metabolism; 6(1):17-24.
View at Publisher | View at Google Scholar - Burke W, Psaty BM., (2007), Personalized medicine in the era of genomics, The Journal of American Medical Association; 298(14):1682-1684.
View at Publisher | View at Google Scholar - Suhad Hafidh, et al., (2023), Insights on Pharmacogenetics and Pharmacogenomics: Advantages and Challenges for Healthcare Professionals. Journal of Medical Research and Health Sciences; 6 (9):27032-2741.
View at Publisher | View at Google Scholar - Evans WE, McLeod HL., (2003), Pharmacogenomics-drug disposition, drug targets, and side effects. The New England Journal of Medicine; 348(6):538-549.
View at Publisher | View at Google Scholar - Bekesho et al., (2016), Pharmacogenetics and Pharmacogenomics: Applications in Drug Development and Therapeutics. EC Pharmacology and Toxicology; 2(4):165-186.
View at Publisher | View at Google Scholar - Kirchheiner J, Brockmoller J, (2005), Clinical consequences of cytochrome P450 2C9 polymorphisms. Clinical Pharmacology; 77(1):1-16.
View at Publisher | View at Google Scholar - Philip Board et al., (1998), Polymorphism of phase II enzymes: identification of new enzymes and polymorphic variants by database analysis. Toxicology Letters; 102(103):149-154.
View at Publisher | View at Google Scholar - Katy Walker et al., (2009), Genetic polymorphism in N-Acetyltransferase (NAT): Population distribution of NAT1 and NAT2 activity Journal of Toxicology and Environmental Health; 12(5-6):440-472.
View at Publisher | View at Google Scholar - Marcuello E, Altes A, Menoyo A, et al., (2004), UGT1A1 gene variations and irinotecan treatment in patients with metastatic colorectal cancer. British Journal of Cancer. PubMed; 91:678-682.
View at Publisher | View at Google Scholar - Federica Sangiuolo et al., (2002), Towards the pharmacogenomics of cystic fibrosis Pharmacogenomics; 3(1):75-87.
View at Publisher | View at Google Scholar - Bradley S Quon, (2015), A new era of personalized medicine for cystic fibrosis – at last! Canadian respiratory journal; 22(5): 257–260
View at Publisher | View at Google Scholar - Vaxillaire M, Froguel P. Monogenic diabetes in the young, pharmacogenetics and relevance to multifactorial forms of type 2 diabetes. Endocrine Reviews; 29(3):254-264.
View at Publisher | View at Google Scholar - Chunmei Huang, (2011), Pharmacogenetics in type 2 diabetes: potential implications for clinical practice. Genome Medicine 2011; 3(76).
View at Publisher | View at Google Scholar - James McGough M.D. et al., (2006), Pharmacogenetics of Methylphenidate Response in Preschoolers With ADHD. Journal of the American Academy of Child & Adolescent Psychiatry; 45(11):1314-1322.
View at Publisher | View at Google Scholar - Brown JT.et al., (2022), The Pharmacogenetic Impact on the Pharmacokinetics of ADHD Medications. Methods in Molecular Biology; 2547:427-436.
View at Publisher | View at Google Scholar - Jesús K Yamamoto-Furusho, (2017), Pharmacogenetics in inflammatory bowel disease: understanding treatment response and personalizing therapeutic strategies. Pharmacogenomics and Personalized Medicine; (10):197-204.
View at Publisher | View at Google Scholar - Marie Pierik, (2006), Pharmacogenetics in inflammatory bowel disease. World Journal of Gastroenterology; 12(23):3657-3667.
View at Publisher | View at Google Scholar - E Tanaka, (2004), Pharmacogenetics of disease-modifying anti-rheumatic drugs. Best Practice & Research Clinical Rheumatology; 18(2):233-247.
View at Publisher | View at Google Scholar - Mohamed H. Babiker-Mohamed, (2024), Pharmacogenetics of therapies in rheumatoid arthritis: An update Best Practice & Research Clinical Rheumatology; 38(4):1521-6942.
View at Publisher | View at Google Scholar - Hull, Larson, A., (2004), Pharmacogenetic Candidate Genes for Melanoma. Pharmacogenomics; 4(6):753-765.
View at Publisher | View at Google Scholar - Goey, A. K., (2019), With Effects of Pharmacogenetic Variants on Vemurafenib-related Toxicities in Patients with Melanoma. Pharmacogenomics; 20(18):1283-1290.
View at Publisher | View at Google Scholar - Kelly Westbrook, (2013), Pharmacogenomics of Breast Cancer Therapy: An Update. Pharmacology and Therapeutics; 139(1):1-11.
View at Publisher | View at Google Scholar - Ram Mohan Ram Kumar, (2024), Enhancing breast cancer treatment through pharmacogenomics: A narrative review, Clinica Chimica Acta; 2024-562
View at Publisher | View at Google Scholar - Altman RB and Klein TE., (2002), Challenges for biomedical informatics and pharmacogenomics. Annual Review of Pharmacology and toxicology; 42:113-133.
View at Publisher | View at Google Scholar

 Clinic
Clinic
